| MRIcro Anatomical Templates |
New: This page describes new MRIcro features and requires MRIcro 1.35 or later.
MRIcro can load anatomical template images. These templates can help you estimate the location of different regions of the brain. Furthermore, neuropsychologists can use these images to examine which areas are commonly damaged in a group of patients. Two example templates are the Brodmann Areas image and the Automated Anatomical Labelling map. However, you can also create your own custom anatomical templates. For more details on the AAL map, you may wish to read:
| Tzourio-Mazoyer N et al. Automated anatomical labelling of activations in spm using a macroscopic anatomical parcellation of the MNI MRI single subject brain. Neuroimage 2002; 15: 273-289. |
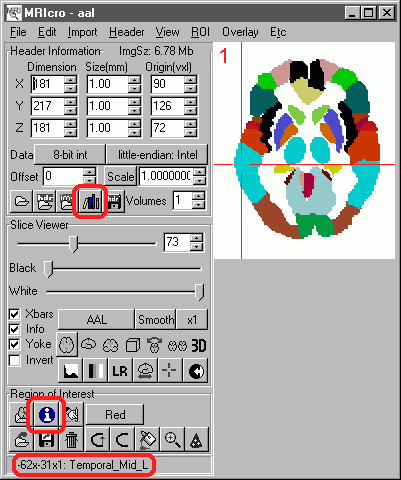
| Using Anatomical images Start MRIcro, and load the desired map (select 'Open image' from the 'File' menu and then choose the map you wish to view, e.g. the AAL map). The desired map should then appear (as shown on the left).Note that when your mouse moves over a region of the brain, the label appears at the bottom-left of the MRIcro window (e.g. in this example the crosshair is over the Temporal_Mid_L). You can also use these anatomical templates to estimate locations on other brains that have been normalized (automatically rotated and stretched to match the overall shape). To do this, start two copies of MRIcro, one viewing the anatomical template image and one showing the individual's MRI scan. When you click on any location in either scan, the corresponding region should appear in the other one, along with the anatomic label (again at the bottom right of the copy of MRIcro displaying the anatomic template). Note that the 'Yoke' button needs to be checked for this to work (it is shown checked in the image on the right). Remember that each individuals brain has a unique set of sulcal folds, so the positions will be approximate. For more details on yoking two images, see my MRIcro tutorial. You can also find out the regions of a brain that have been damaged in an indivdual. After creating a region of interest (ROI), press the 'i'nformation button (shown on the right). This will report the number of voxels damaged (note that you must concurrently load the anatomical template and the desired ROI. A sample report might read:
See my other pages for more details about creating ROIs and lesion analysis. |
 |
You can also create your own anatomical template. The AAL image that is included is a nice example of how a custom template can be created. To create a template you should have:
 |