| The DICOM Standard |
 |
This guide gives is a brief description of the DICOM standard, which is commonly used for the transfer and storage of medical images. |
| An introduction to
the DICOM single-file format The Digital Imaging and Communications in Medicine (DICOM) standard was created by the National Electrical Manufacturers Association (NEMA) to aid the distribution and viewing of medical images, such as CT scans, MRIs, and ultrasound. Part 10 of the standard describes a file format for the distribution of images. This format is an extension of the older NEMA standard. Most people refer to image files which are compliant with Part 10 of the DICOM standard as DICOM format files. A complete copy of the standard (in PDF format) is avaiable for download (drafts of the standard are organized by year). A single DICOM file contains both a header (which stores information about the patient's name, the type of scan, image dimensions, etc), as well as all of the image data (which can contain information in three dimensions). This is different from the popular Analyze format, which stores the image data in one file (*.img) and the header data in another file (*.hdr). Another difference between DICOM and Analyze is that the DICOM image data can be compressed (encapsulated) to reduce the image size. Files can be compressed using lossy or lossless variants of the JPEG format, as well as a lossless Run-Length Encoding format (which is identical to the packed-bits compression found in some TIFF format images). DICOM is the most common standard for receiving scans from a hospital. Neuroimagers and neuropsychologists who wish to use SPM to normalize scans to stereotaxic space will need to convert these files to Analyze format. My freeware MRIcro software will directly convert most DICOM images to and from Analyze format. Eric Nolf's free Medcon and XMedcon software can also convert between Analyze and DICOM. |
 |
|
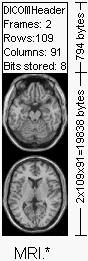
The
DICOM header The Image on the left shows a hypothetical DICOM image file. In this example, the first 794 bytes are used for a DICOM format header, which describes the image dimensions and retains other text information about the scan. The size of this header varies depending on how much header information is stored. Here, the header defines an image which has the dimensions 109x91x2 voxels, with a data resolution of 1 byte per voxel (so the total image size will be 19838). The image data follows the header information (the header and the image data are stored in the same file). Further down, I show a more detailed list of the DICOM header as displayed by my software. Note that DICOM requires a 128-byte preamble (these 128 bytes are usually all set to zero), followed by the letters 'D', 'I', 'C', 'M'. This is followed by the header information, which is organized in 'groups'. For example, the group 0002hex is the file meta information group, and (in the example on the left) contains 3 elements: one defines the group length, one stores the file version and the third stores the transfer syntax. The DICOM elements required depends on the image type, and are listed in Part 3 of the DICOM standard. For example, this image modality is 'MR' (see group:element 0008:0060), so it should have elements to describe the MRI echo time. The absence of this information in this image is a violation of the DICOM standard. In practice, most DICOM format viewers (including MRIcro and ezDICOM) do not check for the presence of most of these elements, extracting only the header information which describes the image size. The NEMA standard preceded DICOM, and the structure is very similar, with many of the same elements. The main difference is that the NEMA format does not have the 128-byte data offset buffer or the lead characters 'DICM'. In addition, NEMA did not explicitly define multi-frame(3D) images, so element 0028,0008 was not present. Of particular importance is group:element 0002:0010. This defines the 'Transfer Syntax Unique Identification' (see the table on the left). This value reports the structure of the image data, revealing whether the data has been compressed. Note that many DICOM viewers can only handle uncompressed raw data. DICOM images can be compressed both by the common lossy JPEG compression scheme (where some high frequency information is lost) as well as a lossless JPEG scheme that is rarely seen outside of medical imaging (this is the original and rare Huffman lossless JPEG, not the more recent and efficient JPEG-LS algorithm). These codes are described in Part 5 of the DICOM standard. A nice introduction to this the transfer syntax is provided at www.barre.nom.fr. Note that as well as reporting the compression technique (if any), the Transfer Syntax UID also reports the byte order for raw data. Different computers store integer values differently, so called 'big endian' and 'little endian' ordering. Consider a 16-bit integer with the value 257: the most significant byte stores the value 01 (=255), while the least significant byte stores the value 02. Some computers would save this value as 01:02, while others will store it as 02:01. Therefore, for data with more than 8-bits per sample, a DICOM viewer may need to swap the byte-order of the data to match the ordering used by your computer. In addition to the Transfer Syntax UID, the image is also specified by the Samples Per Pixel (0028:0002), Photometric Interpretation (0028:0004), the Bits Allocated (0028:0100). For most MRI and CT images, the photometric interpretation is a continuous monochrome (e.g. typically depicted with pixels in grayscale). In DICOM, these monochrome images are given a photometric interpretation of 'MONOCHROME1' (low values=bright, high values=dim) or 'MONOCHROME2' (low values=dark, high values=bright). However, many ultrasound images and medical photographs include color, and these are described by different photometric interpretations (e.g. Palette, RGB, CMYK, YBR, etc). Some colour images (e.g. RGB) store 3-samples per pixel (one each for red, green and blue), while monochrome and paletted images typically store only one sample per image. Each images store 8-bits (256 levels) or 16-bits per sample (65,535 levels), though some scanners save data in 12-bit or 32-bit resolution. So a RGB image that stores 3 samples per pixel at 8-bits per can potentially describe 16 million colours (256 cubed). |
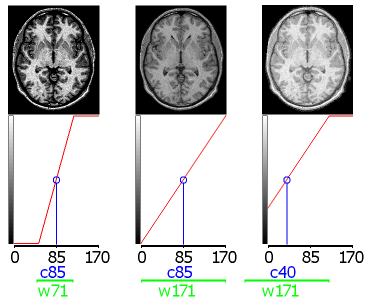
| Window center
and window width (aka brightness and contrast) People familiar with the medical imaging typically talk about the 'window center' and the 'window width' of an image. This is simply a way of describing the 'brightness' and 'contrast' of the image. These values are particularly important for Xray/CT/PET scanners that tend to generate consistently calibrated intensities so you can use a specific C:W pair for every image you see (e.g. 400:2000 might be good for visualising bone, while 50:350 might be a better choice for soft tissue). Note that contrast in MRI scanners is relative, and so a C:W pair that works well for one protocol will probably be useless with a different protocol or on a different scanner. The figure on the right illustrates the concept of changes to'window center' and 'window width'. Along the top row you can see three views of the same image with different C:W settings. The bottom row illustrates the colour mapping for each image (with the vertical axis of the graph showing rendered brightness and the horizontal axis showing the image intensity). Consider this image with intensities ranging from 0 to 170. A good starting estimate for this image might be a center of 85 (mean intensity) and width of 171 (range of values), as shown in the middle panel. Reducing the width to 71 would increase the contrast (left panel). On the other hand, keeping a width of 171 but reducing the center to 40 would make the whole image appear brighter. |
 |
Links to DICOM servers and clients
Links to DICOM image conversion
 |