 |
Features:
- Converts medical images to SPM
friendly Analyze format.
- View Analyze format images (big or
little endian).
- Create Analyze format headers (big
or little endian).
- Create 3D regions of interest
(with computed volume & intensity).
- Overlap multiple regions of
interest.
- Rotate images to match SPM
template images.
- Export images to BMP, JPEG, PNG or
TIF format.
- Yoked images: linked viewing of
multiple images (e.g. view same coordinates of
PET and MRI scans).
|
Index
- Introduction
- Installation
- Loading images and the header
information panel
- Slice viewer panel
- Region of interest panel
- Rotation and clipping panel
- Overlays [Statistical Maps]
- Selecting Coordinates
- Other commands
- Viewing medical images
- Converting medical images to Analyze
format
- Uninstalling
- Technical details
Introduction
Return to Index
MRIcro allows Windows and Linux computers view medical images.
It is a standalone program, but includes tools to complement SPM
(software that allows neuroimagers to analyse MRI, fMRI and PET
images). MRIcro allows efficient viewing and exporting of brain
images. In addition, it allows neuropsychologists to identify
regions of interest (ROIs, e.g. lesions). MRIcro can create
Analyze format headers for exporting brain images to other
platforms.
Users familiar with other Windows programs will find that this
software is fairly straightforward to use. Resting the mouse
cursor over a button will cause a text hint to appear over the
button. As a last resort, I have included this brief manual that
describes the basic features.
A tutorial with a step by step guide of how to use MRIcro with
SPM is available.
Installation
Return
to Index 
Note: MRIcro supports the Analyze file format. I strongly suggest using MRIcron.
MRIcron supports the NIfTI format and has many new features. The only reason to use MRIcro instead of MRIcron is if you are familiar with this software or if you have a computer with limited memory (<256 Mb).
This section describes how to install MRIcro on a computer
with the Windows operating system. There is a separate web page
that describes the installation of the Linux
version of MRIcro.
- With your computer connected to the web, download the 5.9
Mb installer program, choose a mirror that is close to
you:
- Double click on the "mrinstall" icon. The
installer will give you the option to install the
manuals, a sample MRI image and other files. By default,
the files will be installed in "C:\Program
Files\MRIcro". Note: with Windows 2000/NT,
only adminstrators can copy files into the "Program
Files" folder. If you are using 2000 or NT, either
log in as an administrator or choose a different folder
to install the files (e.g.
"C:\username\mricro").
- To run MRIcro, click on the "Start" menu,
select "Programs", point to the
"MRIcro" folder and click on the
"MRIcro" icon.
Loading
images and the header information panel
Return to Index
MRIcro can view various medical image formats, including the
Analyze format used by SPM. Analyze format images have two
components: the image file (*.img) that contains the raw image
data and a header file (*.hdr) that describes the image
dimensions, data format and comments. MRIcro's header information
panel displays the header file's information and includes a
series of buttons that allow you to open and view headers.
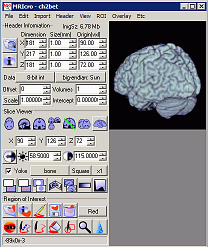
 |
Figure 1. The header information panel. |
The 'open Analyze format hdr/img pair' button (it looks like
an open folder) allows you to view an Analyze or NIfTI format image. When
you press this button, a dialog will appear that allows you to
select a header file to open. MRIcro will then attempt to open an
image file of with the same name (e.g. if you select a header
'C:\17.hdr', MRIcro will attempt to open the image file
'C:\17.img').
The 'show extended header information' button (it looks like a
'i'nformation icon) will open a window showing additional information
about the currently open header (e.g. Scan comments, patient ID,
etc).
Slice
viewer panel Return to Index
The slice viewer panel allows you to select the appearance of
the loaded image file. The slider selects which slice is
displayed (alternately, you can select the slice in the edit box
located directly to the right of the slider, or pressing Fn1/Fn2
to view the next successively lower/higher slice; if your mouse has a scroll wheel you can roll the wheel to change slice).
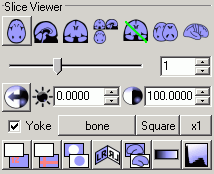 |
Figure 2. The slice viewer panel. When viewing
axial, sagittal or coronal views, a slider appears that
allows you to set which slice is displayed (illustrated).
When projection views are displayed, you can set the X, Y
and Z coordinates independently by adjusting the three
edit boxes. |
The brightness and control the contrast of the image by
changing the values located next to the sun (brightness) and
contrast images. These numbers describe the window
center and window width of the display. The window centre
refers to the image intensity that will be displayed as a
medium-gray and the window width describes the range between
bright white and full black. Clicking the 'auto contrast' button
(showing a black arrow on
a mostly white circle) sets 1% of the image to be maximum black
and 1% to be maximum white. The autobalance works well for MRI
scans, but often is not appropriate for CT scans, where the bones
appear much brighter than the background and brain tissue. For CT
scans, you can often get good settings by choosing one of the
'Contrast Preset [CT]' in the 'View' window. For example, the
'Bone' preset sets the window center to be 400 and the width to
2000, and is often useful for highlighting bone in CT scans.
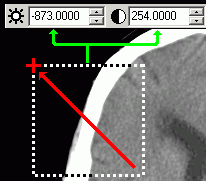 |
Figure 3. You can adjust the contrast for a
specific region by dragging over the area of the image
while depressing the RIGHT mouse button. The brightness
(window center) and contrast (window width) values are
adjusted to scale the selected area. Note that this will
not work if you have selected any of the
region-of-interest drawing tools (press Fn10 to deselect
drawing tools). |
| Figure 4. This histogram shows the image
intensity profile for the image 'IconT1'. Histograms can
also be created for user defined regions of interest (see
the ROI section for more details).
Note the Threshold Mn value is listed, this can be used
to set the image intensity scale of a target image to
match an SPM template (see the tutorial
for more details). |
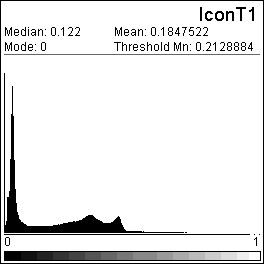 |
The histogram button (located at the bottom-left of the panel)
displays the voxel brightness distribution in the currently open
image (see figure, above).
Six icons appear along the top of the Slice viewer panel
that allow you to select which view of the image is displayed:
axial, sagittal, coronal or projection (axial, sagittal and
coronal simultaneously), free rotate, multislice and 3D
rendering. If the image does not correspond with the selected
button, the image was not saved in axial format. In the
projection view, three edit boxes appear at the top of the slice
viewer, allowing you to independently set the X, Y and Z
coordinates you wish to view (alternately, click on the image to
jump to the mouse coordinates).
If you are viewing more than one image simultaneously (having
launched MRIcro more than once) you can 'yoke' the views, so that
the same view is mirrored for each image (for this feature to
work, make sure that the 'yoke' check box is selected). Yoking
will try to match the slices based on the origin and size
coordinates. This allows you to compare normalisation between an
image and its template, as well as localising structures on a PET
scan by comparing the PET scan with its coregistered T1 MRI scan.
To see a series of transverse or coronal slices simultaneously
press the 'multislice' button (it has an icon showing three axial
slices next to a sagittal slice). In order to function, an image
must be loaded. The slices can be selected in the 'options'
window, as described in other commands
section.
Selecting the free rotate button causes a new window titled
'Free Rotate' to appear (see the figure below). This window
allows you to select which oblique section you wish to view. You
can independently set the yaw, roll, and pitch of the scan.
Furthermore, you can select which slice you wish to view. The
'pivot' settings allow you to set the axis for the image
rotations. You can also 'nudge' the image (center the image in
the frame). Finally, you can set new image dimensions. Custom
views can be saved as graphic pictures (using the 'file/save as
picture' command), printed, or press the 'Save' button at the
bottom of the Free Rotate window to create a new 8-bit Analyze
format image described by your rotations. Note that the free
rotate tools mix viewer centered and object centered coordinates,
which can become somewhat disorienting. Also, when free rotating
multiple Regions of interest with the 'interpolate' box checked,
you may see a slight white halo around the regions.
You can generate 3D volume renderings by pressing the '3D'
button (showing a sagittal rendering of the cortex). This feature is described in more detail on my volume rendering page.
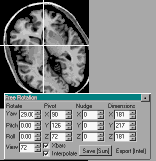 |
Figure 5. The free rotation view allows you to
create a custom view. |
The Mirror button (it displays the letter 'LR' in either
standard or mirrored text, depending on whether mirroring is
selected) allows you to flip the image's left and right. If you
select mirroring, a message will appear reminding you that it is
best not to edit regions of interest while in the mirror mode (as
the ROI will be stored in a different orientation than the
original image).
A drop-down box selects the color lookup table (LUT) used. By
default, MRIcro can show images in the 'black & white' or
'hot metal' color schemes. However, MRIcro can also read Osiris
format LUTs, which allow you to view images in different color
schemes. MRIcro ships with several LUTs from the Osiris distribution, and you
can create
your own custom LUTs. If MRIcro finds any *.LUT files in its
directory, the drop-down box will list the file names. Select the
color scheme you want from the drop down box.
A drop down box allows you to select how MRIcro displays
images with voxels of unequal dimensions (e.g. an image slice
where voxels are 2mm in theX dimension but 1mm in the Y
dimension). First, images can be viewed in the 'square' mode,
where all voxels are displayed as being square, irregardless of
interslice dimensions. Second, you can view images 'stretch'ed,
where the size of voxels is proportional to the interslice
dimensions, using a nearest neighbor approximation (though
projection and multislice views are NOT scaled). Finally, you can
view images as 'smooth'ed: voxel size is proportional and uses a
nearest neighbor approximation (this creates 24-bit images,
projection views are shown correctly scaled).
Use the zoom factor box (located at the lower right of the
slice viewer panel) to set the image scale, MRIcro allows you to
view images in x1-x6 scale. Depending on the size and scale of
your images, you may want to adjust the size of the MRIcro window
(by dragging the lower right corner of the viewer).
Region
of interest panel Return to Index
MRIcro allows you to draw three-dimensional regions of
interest (ROIs). This is useful for illustrating regions of the
brain that have sustained damage. In addition, the volume of the
ROI is computed. Moreover, ROIs from different individuals can be
overlapped (on brain images that have been normalised to the same
template) allowing neuropsychologists to assess common areas of
damage.
ROIs can be saved to disk for future reference. All ROIs are
drawn on top of the MRI image, rather than directly on it. This
means that the brain images can be viewed with or without
corresponding ROIs.
MRIcro provides a number of tools for creating and viewing
ROIs. These tools are displayed as a series of buttons in the
'Region of interest' panel. These buttons will be described
sequentially from left to right.
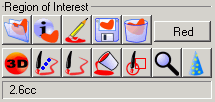 |
Figure 6. The Region of
Interest panel contains tools for marking lesion
locations or specific areas. When you load a ROI, the
volume is displayed in the bottom left corner, here 2.6
cubic centimeters. |
The 'open roi' button (it looks like an open folder with a
ROI) allows you to select ROI[s] to open. If you want to open
multiple ROIs simultaneously, depress the control key as you
select the ROI names.
The ROI information button (it displays an 'i'nformation icon)
displays the mean image intensity of a region of interest as well
as generating an image intensity histogram (similar to the
histogram depicted in the slice viewer section).
The 'save ROI' button (it has an icon of a disk with a ROI
superimposed on it) allows you to save a ROI that you have
created to disk.
Use the 'delete ROI on this slice' button (it shows a pencil
erasing information) to remove the ROI only from the slice you
are currently viewing. This button is useful if you make a
mistake outlining or filling a ROI that you are drawing.
In order to remove all current ROI[s] from memory, click the
'delete entire ROI' button (it looks like a waste basket). If you
have opened multiple ROIs, you will need to select this before
creating a new ROI (you can only write to one ROI at a time).
The 'save ROI' button (it has an icon of a disk with a ROI superimposed on it) allows you to save a ROI that you have created to disk.
The 'Delete entire ROI' button (it has an icon of a wastebasket with a ROI superimposed on it) allows you to close a ROI.
Next, there is a small drop down box that defines the colour
that single ROIs will be drawn in (when viewing multiple ROIs
simultaneously a rainbow colour set is automatically used). The
choices are Red, Green, Blue, White or Black. This allows you to
choose a salient colour. You can set the ROI to black or white
before printing the image to a black and white printer.
After loading multiple ROIs, you can select the 'ROI density
colorbar' item from the 'ROI' menu to draw graphic of the overlap
between ROIs. The colours indicate the number of overlapping
ROIs. The leftmost (dark violet) colour indicates the index for a
single ROI, while the rightmost (bright red) colour shows the
index for all the ROIs overlapping. This feature is demonstrated
in the figure below:
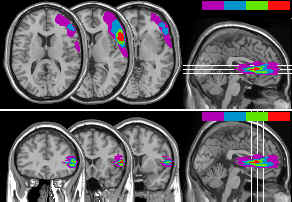 |
Figure 7. These figures show MRI multislices
with four overlapping ROIs. The colour bar in the upper
right corners indicates the ROI density (dark violet for
one ROI, bright red for four overlapping ROIs). These
figures also demonstrates the option window's 'show right
hemisphere' function -where successive slices overlap
each other. Multislices can be generated either for
transverse slices (top panel) or coronal slices (bottom
panel). |
Select one of the pens for creating ROI outlines. There are
two pens: the 'closed' pen (shortcut key Fn6) automatically
closes any outline you have drawn. The 'open' pen (shortcut key
Fn7) does not automatically close the outline. The 'fill button'
(paint can icon, shortcut Fn8) can be used to fill a ROI that has
been outlined by the pen (alternative: right click with the pen
selected). Before beginning to create a new ROI you should load
the image, clear any previous ROIs (using the 'save ROI' or
'delete entire ROI' buttons described above), and set the view to
transverse, coronal or sagittal slices (you can not draw ROIs on
the free rotate, projection and multiple views). Holding the
shift key down while using a pen or fill will erase ROIs in the
designated region.
In practice, ROIs can be drawn rapidly by selecting the closed
pen (shortcut key Fn6) and then using Fn1 and Fn2 to move up and
down to the desired slices. Using the pen with the shift key down
is useful for trimming unwanted edges from a ROI. Use the left
mouse button to outline the ROI, and then move to the centre of
the ROI and right-click to fill the region. Once the ROI is
drawn, use the 'Save ROI' button to store the ROI. Note that the
ROI volume is listed in the Region of interest panel (computed in
either cc or voxels, updated when you change slices or save the
ROI).
The wizard's cap icon allows you to temporarily hide a region
of interest (shortcut key Fn9). By rapidly hiding and showing the
ROI, you can see whether the ROI you have drawn correctly maps
the region you are interested in.
When the mouse moves over an image, two useful bits of
information are displayed immediately below the region of
interest panel. Text on the far left side shows the the mouse
position in Talaraich space, followed by the image intensity
immediately beneath the mouse.
Rotation
and clipping panel Return to Index
The file menu contains a command labelled 'Save as...'. When
this command is selected, a floating panel appears in the lower
left corner of program's window. The rotation and clipping panel
can be used to prepare image files for normalisation
(coregistering an image to a standard template) with SPM.
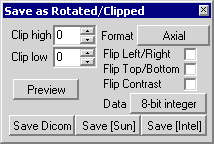 |
Figure 8. The rotation and clipping panel. |
The clip top and clip bottom fields allow you to choose the
number of slices that you want to shave off of the top or bottom
of a scan (for example, trimming scans that include too much
spinal cord to avoid problems with SPM normalisations). The
'Format' list box allows you to convert coronal or sagittal
Analyze format images to axial format (this is the natural
preference for SPM). To determine the original format of your
image, click on the 'axial [flat] view' button in the slice
viewer panel, this will display the scan in its native format
(e.g. if the flat format displays a sagittal image, the file is
in sagittal format). The 'flip left/right' checkbox mirrors the
image.
You can also set the 'data' type for the saved image. Usually
it is wise to save the data in the same format as the source
image. Down-sampling an image (e.g. saving a 16-bit integer file
as an 8-bit data file) will save disk space (potentially at the
expense of image intensity quality). Up-sampling an image (e.g.
saving a 16-bit file as a 32-bit file) can be useful for when
other programs require data of a specific type. If you have
adjusted the contrast/brightness settings in the slice viewer
panel, you will be asked if you wish to clip the brightest and/or
darkest voxels, allowing you to customise the contrast of the
output.
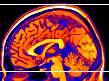 |
Figure 9. MRIcro automatically displays the
amount of clipping selected, in this example the top five
and bottom ten slices are about to be clipped. This
figure also shows the 'Hot Metal' color lookup table. |
Before saving an image that you wish to rotate, you can check
your settings by pressing the 'preview' button. The preview will
show you two slices of how the image will appear after being
rotated. If your settings are correct, the preview should show
two transverse slices (with the left slice being more ventral
than the right slice). Make sure to check that the left/right
mirroring is correct.
When you have selected the desired clipping and check box
options, press one of the three file save buttons that are
located at the bottom of the window. The buttons 'Save [Sun]' and
'Save [Intel]' will save the files as Analyze format images.
SPMwin, SPM99, SPM2 and MRIcro can all read either big or
little-endian Analyze files, while SPM96 requires the images to
be in the same format as the machine used. For more information,
see the technical section.If the
original image is a multi-volume image (i.e. the img file
contains multiple MRI scans, which are all the same dimension),
MRIcro will rotate each volume and save it as a separate Analyze
format header/image (this is useful, as SPM can not read multiple
volume files). The 'Save Dicom' button will save the image as a
DICOM format image (as described in the DICOM
section of this manual). Note that the DICOM format image
will either be in 8-bit integer (if that is the data type of the
source image) or 16-bit integer format irregardless of the data
type seleted.
Overlays
[Statistical Maps] Return to Index
The 'Overlay' menu allows you to select an image which is
superimposed on top of another image. This is useful for
displaying functional statistical maps (generated by SPM from
PET, fMRI or SPECT data). Overlays can also be used to check the
coregistration of two images. To display an overlay, you should
first choose the primary image you want to use (e.g. use the
'Open' command from the file menu to select your anatomical
image). Next, use the 'Load overlay' command in the Overlay menu
to select the image you wish to superimpose. The overlay does NOT
need to be the resliced to the same dimensions as the anatomical
image: MRIcro will correctly reslice the data (as long as the
image dimensions and origin are correctly specified for each
image). The overlay menu also allows you to select the color of
the overlay and whether the overlay is shown as opaque or
transparent. The images below show overlays of functional data
(left) and the use of the overlay function to check
normalization. For more details, visit my overlay and volume rendering
pages.
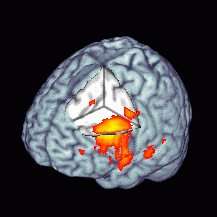 |
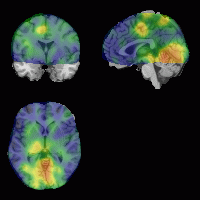
Left: functional results can be overlayed.
Above: you can use overlays to check alignment after
normalization. |
Selecting
coordinates Return to Index
The 'Select MNI/Talairach Coordinates' command in the 'View'
menu allows you to specify which region of the brain you want to
view. With the slice viewer panel, you
choose which slice in the image you wish to view. In contrast,
the coordinates command allows you to type in the steotactic
coordinates used by neuroimagers. This command only makes sense
if your image has been normalized (stretched, rotated and
centered to match a standard neurological frame of reference).
There are two popular frames of reference for brain imaging: MNI
space (used by SPM) and Talairach space (used in the atlases of
Talairach and Torneaux). A check box in the coordinates window
allows you to set which frame of reference you wish to use (the
Talairach coordinates are estimated used Matthew Brett's handy mni2tal
and tal2mni routines). These two methods are illustrated
below. When looking at an image from the Talairach and Tournoux
atlas (left) you should make sure the 'tal2mni' box is checked
(shown in red). On the other hand, when looking at SPM results,
make sure this feature is switched off.
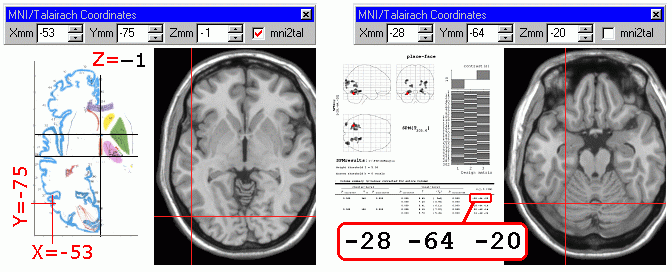
Other
commands Return to Index
The 'File' menu contains the command 'save as picture', which
allows you to save the currently displayed image as a 2D graphic
image. Supported formats include the popular BMP, JPEG, PNG and
TIF formats. The File menu's 'Print' item allows you to print the
currently displayed image.
The ROI menu contains commands that allow you to export ROIs
as big-endian (Sun) or little-endian (Intel) Analyze format
images. These commands save the currently displayed region of
interest as an 8-bit Analyze format image, which can be used in
SPM as a mask (allowing sensible normalization of brain images
that have large lesions). Note that the only values on the mask
image are 0 (unmasked) and 100 (masked), and that the mask's
image intensity scale value is set to 0.01 (so from SPM's point
of view, the Mask has voxels of the values 0 and 1).
The ROI menu's 'Export Image as ROI' converts an Analyze
format image in MRIcro's custom ROI format. Note that the ROI
format is binary -the ROI does not store intensity information.
Load the image you wish to convert before selecting this command.
When you select this item, a window appears that allows you to
set the intensity ROI's threshold. As you adjust the threshold,
the image will will preview the portions of the image will be
included in the ROI. When you are happy with the selection, press
the 'Save as ROI' button to create the new ROI.
In addition, you can transfer a region of interest between two
images that have different dimensions. One example of the utility
of this command is identifying cortical regions on a PET scan.
Using the 'File/Transfer ROI' command you can create a ROI on the
individuals T1 anatomical MRI scan and then copy the ROI to the
corresponding PET scan(s).
The 'Etc' menu's command 'Options' allows you to adjust a
number of MRIcro's settings. First of all, you can select which
slices will appear when the 'multislice' button is pressed. There
are twelve data fields, allowing up to twelve slices to be viewed
simultaneously. Fields that are set to zero will not be
presented, allowing you to show fewer than twelve slices. You can
also select whether the multislice button will generate coronal
or transverse images. The 'defaults' button at the bottom of the
window will select a common set of slices useful for viewing
images that have been normalised to SPM's T1 template. The
'include sagittal' check box selects whether or not a sagittal
view will appear when the 'multislice' button is pressed. The
option window also includes a number of additional check boxes
that describe how the multislice view will be presented. A few of
the less intuitive options are shown in the figures below. Once
you have selected the desired options, you can press 'OK' to save
your selection, or 'Cancel' to ignore your changes. A number of
these options are described in the tutorial, and some are
illustrated in the figures below.
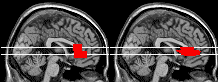 |
Figure 10. If the option window's 'Cumulative
ROI boxed [CT]' check is selected, the sagittal view will
extrapolate regions from the selected slices (left
panel). In this case, ROIs only need to be drawn on the
slices that will be viewed. Otherwise, the sagittal view
will show ROIs for each slice independently. |
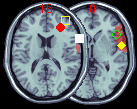
| Figure 11. Right clicking the multislice
button allows you to superimpose hotspots onto an image.
The hotspots are defined in a text file, which includes
the X Y Z coordinates and the colour/shape of the figure
to be superimposed. This can be useful for comparing a
neurological patient's lesion to hotspots from fMRI/PET
studies. In this example, a number of hotspots have been
drawn on top of a frontal lobe ROI. For more details, see
the MRIcro
FAQ. This picture also illustrates the 'translusent
ROI' feature. |
 |
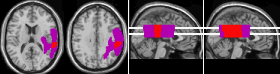 |
Figure 12. The left panel shows axial slices
of two ROIs -one is primarily subcortical and the other
is primariily cortical. When the option window's
'Cumulative only true overlap' is selected, the sagittal
view will show ROI densities based on the maximum true
density of ROIs (middle panel). If this option is not
selected, the sagittal view ROI will sum all ROIs,
independent of their location on the X axis (right
panel). Note that this will sum ROIs irrespective of
whether they are both cortical/subcortical or even in the
same hemisphere. |
Viewing
medical images Return to Index
In addition to Analyze format files, MRIcro can also view a
wide range of standard medical image formats. A full list of
these formats is listed in the Technical
Details section of this manual. MRIcro will automatically
detect the format of your image regardless of whether it is
Analyze, DICOM, etc. format. The easiest way to open a medical
image is to simplay drag and drop its icon onto MRIcro.
Alternatively, select the 'Open foreign...' command from the
'Import' menu and select your image. One exception to this rule
is ECAT images: MRIcro will not detect ECAT6 files and can only
automatically view some ECAT7 files. For ECAT files, one should
first convert the file to Analyze format (as described in the
next section).
Converting
medical images to Analyze format
Return to Index
SPM, Analyze, mri3dX and MRIcro all use the Analyze format as
their native image format. However, many scanners save images to DICOM
or proprietary file formats. The technical
section describes the formats that MRIcro and other programs
can convert to Analyze format.
When you convert medical images to Analyze format, you can
select a series of 2D images that will be
stacked and saved as a single unified 3D Analyze format file.
Selecting 'Convert foreign to Analyze' from the 'Import' menu
will create a new window that allows you to describe your images.
My tutorial describes this
process in detail. ECAT images can be converted selecting the
'Convert ECAT to Analyze' command from the import menu. ECAT
images can be saved as the raw data or as scaled data (taking
into account the calibration and scaling factors) - the
conversion format can be selected in by choosing 'Etc/Options'
and setting the 'ECAT convert' values. Finally, you can convert
SPMwin headers to SPM headers using the 'Import/SPMwin VHD to
Analyze' command.
MRIcro can also convert Analyze format images to the DICOM
format. The command 'Save as...' allows you to save an convert an
image to DICOM. The *.img file that is created when you press
'Save DICOM' will be in the DICOM format. The image may not
strictly obey the guidelines of the DICOM standard. However, the
images should be compatible with most DICOM image viewers. The
DICOM file will be either 8-bit (if the source file uses 8-bits
per voxel) or 16-bit (if the source file uses 16-bits or greater
per voxel). The DICOM file that is created is 'anonymized' -and
will contain no private details from the Analyze header (e.g.
patient name, scan date, etc).
Uninstalling
Return to Index
To uninstall MRIcro for Windows, select the
"Add/Remove Programs" control panel (to access the
control panel, select StartMenu/Settings/ControlPanel). Simply
select "MRIcro [remove only]" from the "Add/Remove
Programs" control panel and press "Add/Remove".
In order to remove MRIcro for Linux from your computer,
delete the folder "/usr/local/bin/mricro". You will
need root permissions to do this.
MRIcro normally remembers the recent files you used, your
preferred views, and other information. If you want to reset
MRIcro to the default settings, run the program and select the
'Uninstall' command from the 'Etc' menu. Then quit MRIcro and
relaunching the program.
Technical
Details Return to Index
- Referencing. Scientific publications can refer to
Rorden, C., Brett, M. (2000).
Stereotaxic display of brain lesions. Behavioural
Neurology, 12, 191-200.
- License. The compiled MRIcro application is provided as freeware under the BSD license. Portions of the source code are available from the ezDICOM project at sourceforge.
- Chris Rorden's MRIcro, copyright 1999-2005, all rights reserved.
- Redistribution and use in binary forms, with or without modification, are permitted provided inclusion of the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution.
- Neither the name of the copyright owner nor the name of this project (MRIcro) may be used to endorse or promote products derived from this software without specific prior written permission.
- THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDER "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT OWNER BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
- Trouble shooting.
- If MRIcro initially shows an image correctly but
does not update the display when the slice is
changed, turn off the 'graphics acceleration'
option found in the 'Etc' menu's 'option' window
(graphics acceleration greatly speeds image
display, but is not compatible with all graphics
cards).
- When loading multiple regions of interest (in
order to show mutual regions of overlap) some
Windows NT users may find that not all of the
ROIs are loaded. Some versions of NT will only
report the first 255 characters of file names. A
quick workaround is to shorten the length of the
file names (e.g. instead of 'roiforfile1.roi',
name the file '1.roi'). To check the number of
ROIs that are loaded, choose 'ROI density
colorbar' ('ROI' menu), the number of colours
displayed shows the number of ROIs loaded.
- In addition to this manual, the MRIcro FAQ,
volume rendering page
and the tutorial
resolve most questions. For interactive support,
you can also visit the JISCmail
MRIcro List. Please realise that this
software is downloaded more than 35,000 times per
year. I am happy to help, but it is easy to feel
overwhelmed by questions. You can get help from
the MRIcro users list, either using the JISCmail
web site.
- Foreign language manuals: Tsutomu
Soma has kindly created a Japanese
translation of this manual.
- Usage. MRIcro is designed for research use, rather
than clinical use. The author accepts no liability for
the use of this software. Please use this tool sensibly.
- Memory. MRIcro requires a large amount of Random
Access Memory (RAM) in order to view Analyze format
images. Images with a data type of 8-bit integer will
require one byte per image voxel. Images of 16-bit
integer data type require 3 bytes per voxel (one byte for
the screen buffer and two bytes for the data). Images
with any other data type (e.g. 32-bit integer) require
five bytes per voxel (one byte for the screen image
buffer and four bytes for a real-number array that allows
contrast optimization). Editing or viewing ROIs will
require an additional byte per voxel. For example,
viewing the SPM template (91x109x91 voxels, 8 bit integer
data type) will require .86 Mb of RAM (1.72Mb when
viewing or editing a ROI). The table below lists the
bytes per voxel (bpv) for images of different data types.
In addition, the table lists the RAM required to open a
256x256x124 voxel image of each data type (in megabytes).
The 'Etc' menu's 'About' command will display the RAM
currently being used.
| Data Type |
RAM when loading |
RAM when viewing |
RAM with ROI |
| 8-bit |
1 bpv (7.75 Mb) |
1 bpv (7.75 Mb) |
2 bpv (15.5 Mb) |
| 16-bit |
3 bpv (23.25 Mb) |
3 bpv (23.25 Mb) |
4 bpv (31 Mb) |
| Other |
32-bit: 5 bpv (38.75 Mb)
64-bit: 12 bpv (93.00 Mb) |
5 bpv (38.75 Mb) |
6 bpv (46.5 Mb) |
- Y2K. MRIcro is year 2000 compliant.
- Monitor depth. MRIcro uses a palette of 256
colours for drawing brain images. Because Windows
reserves a number of colours, your display should be set
to a 16-bit (high colour) 24-bit or 32-bit (true colour)
colour palette. If your computer can not support more
than 256 colours, you should switch off the 'graphics
acceleration' check box in the options window (select
'options' from the Etc menu). To find out the current
setting, open the 'display ' control panel and select the
'settings' tab. Using a colour palette of less than 16
bits (e.g. 256 colours) will limit the levels of gray
that MRIcro can display. MRIcro renders images in 255
grays (reserving one colour for the region of interest).
Fewer gray levels are used when viewing multiple regions
of interest.
- Big-endian and little-endian data. Different
computers use different formats for ordering numeric
values that require more than one byte. With
little-endian processors such as Intel and compatibles
the bytes with a higher address are most significant. In
contrast, big-endian Sun, Silicon Graphics, Motorola and
HP processors use the opposite byte order. Alpha and
PowerPC processors can use either depending on a compiler
switch. This means that Analyze format headers and some
Analyze format data files (those with more than one byte
per voxel, e.g. data type greater than 8-bit integer) may
be different between different computers. MRIcro, SPMwin,
SPM99, and SPM2 automatically detect the endian-ness of
the headers files. When using MRIcro to create header
files, you should save them to the same format as the
image file.
- Region of Interest. ROIs are saved as
'filename.ROI'. This is a proprietary binary format that
uses run-length encoding (a simple compression algorithm)
to store a relatively compact image of the region of
interest.
- Credits. MRIcro was programmed using Delphi
(object-oriented Pascal). The sophisticated sliders and
real-number edit boxes are components from the free RXlib
component collection. SPM guru Matthew Brett suggested
and helped test a number of useful enhancements (Matthew
has written a number of great web pages that describe how
SPM works, which are available at www.mrc-
cbu.cam.ac.uk/Imaging/). Tom
Womack added the SSE support for faster viewing of 32
and 64-bit images and improved the rendering techniques.
Chris Rorden wrote MRIcro while working on a project
grant from The
Wellcome Trust.
- Header information used by SPM. During loading and
saving of header files, MRIcro will show an alert message
if it detects an error that might cause SPM difficulty.
SPM requires the header to accurately describe the image
file: the image dimensions, data type and byte offset all
need to be correct. In addition, SPM will use the origin
information to select the voxel deemed to be the 'centre'
of the image, which in normalised images is the centre of
the anterior commissure. If the origin is set to 0,0,0,
then SPM will assume that the centre has not been set,
which usually works pretty well. SPM also uses the
'scale' factor (the last item displayed in the extended
header information window) to compute the true image
intensity. For more information on these topics, search
the SPM
archives. To learn more about the Analyze file
format, view the Mayo
Clinic's extensive guide (www.mayo.edu/bir) or read Medical
Image Format FAQ . For further details on SPM's
specific and nonstandard usage of the Analyze format,
visit the SPM
home or www.mrc-cbu.cam.ac.uk/Imaging/.
- Supported 2D picture formats. MRIcro can open,
save and print BMP, JPEG, PNG and TIF images. Free
viewers such as IrfanView can batch convert images to
other formats such as GIF. JPG images are very compact,
but some of the image quality is lost. BMP and TIF retain
the image quality, but the files are large. PNG (portable
network graphics) format is both lossless and compressed,
combining the best features of TIF and JPG. A number of
PNG viewers are available to support virtually every
major operating system, see www.libpng.org/pub/png/pngapvw.html
for details. My web graphics
page describes the relative merits of these formats.
- Converting images to Analyze
format.
Note: the programs listed below support the Analyze file format. I strongly suggest converting images to the NifTI file format - this extension of the Analyze format stores inportant information. For NIfTI converters please see my dcm2nii webpage
:
| Viewer |
Formats, Notes |
| MRIcro
[Windows, Linux] |
DICOM (uncompressed and
compressed), AFNI (.head), 8bit BMP (.bmp),
BioRad PIC, CTI ECAT6/7, Elscint, Freesurfer, GE
(LX, Genesis, 4.X, 5.X, compressed), Interfile,
NEMA, PGM, PPM, Picker CT, Philips (.PAR/.REC),
PovRay density (.DF3), Siemens (Magnetom Vision,
Somatom, Somatom Plus), Space (.vol), SPMwin
(.vhd), 8bit TIFF (uncompressed), Vista (.v), VFF
(.vff), VoxBo (.cub), Zeiss LSM 510, raw
|
| Bru2Anz
[Windows] |
Bruker Paravision |
| ImageJ
with Analyze
plugin installed [Java:
Windows, Mac, Unix] |
DICOM (uncompressed), BMP, TIFF,
JPEG, GIF, Raw
- Choose File/Open to open your images (if
you want to stack multiple images, place
them alone in a folder and use
File/Import/AllAsStack).
- If your image is 24-bit RGB (e.g. most
JPEGs), you need to convert the image to
grayscale. First, you may want to
optimize the contrast using the functions
in the Process menu. Then use the
Image/Type/8bitGreyscale function.
- Select Plugins/AnalyzeWriter to convert
your images to Analyze format.
- If the color-intensity of the new Analyze
image is inverted, use Plugins/Inverter
before using Plugins/AnalyzeWriter.
|
| XMedCon
[Windows, Unix] |
DICOM (uncompressed), GIF, ECAT6,
Interfile, NEMA |
| Mosaic
to Analyze [Matlab:
Windows, Unix, Mac] |
Siemens Magnetom Vision |
| Syngo
Converter [Matlab:
Windows, Unix, Mac] |
Siemens DICOM (uncompressed) |
| exp2ana3d.m
(for 256x256 images), exp2ana3dt.m
(for 64x64 EPI)[Matlab+SPM:
Windows, Unix] |
Giuseppe Pagnoni's Matlab script
for converting Philips .PAR/.REC export files. |
| ge2spm
[Matlab: Windows,
Unix] |
GE LX, GE 4.X, GE 5.X format |
| stim2analyze
volume_mri_convert [Unix] |
GE Genesis |
| Dicom_Toolkit
[IDL: Windows, Unix,
Mac] |
DICOM |
| VisionToSPM
[Matlab: Windows,
Unix, Mac] |
Siemens Vision |
| ge2spm
[Unix] |
GE Genesis |
| ana2mnc
[ Perl script: Windows,
Unix, Macintosh] |
MINC, Bruker Paravision, GE 4.x,
GE 5.x, DICOM (uncompressed), Scanditronix, ECAT,
Siemens Magnetom, Phillips |
| MINC
to Analyze [Matlab:
Windows, Unix, Mac] |
MINC |
| Image
Converter [Windows] |
Siemens System 7, Shimadzu
HeadTome IV, Hamamatsu Photonics SHR2000, GEMS
2048-15B |
| SPM2
[Matlab: Windows,
Unix, Mac] |
DICOM [Requires Matlab 6.0 or later, only
some DICOM variations, select 'DICOM' from the
'Toolboxes' menu] |
| IMGCON
[Windows, Unix] |
BMP, GIF, PNG, TIF, JPEG, PGM, MIT, EPS, raw |
| MRIConvert
[Windows] |
DICOM (uncompressed) |
| FormatConvert
[Unix] |
Pittsburgh 1.0, Afni, GE (I*.nnn files, E*.MR
files), BrainVoyager (VMR, VTC, STC, VMP) |
| scanSTAT
[Macintosh] |
GE (.MR), ANMR (.irp/.img), MGH |
- Unsupported 3D formats. Please see my
dedicated page describing importing unsupported image
formats using MRIcro. If you a working with raw data,
or a medical format which is not listed in the table
above, you can try this desperate measure to convert the
image to Analyze format:
- First, give the raw data file name the extension
'.img'.
- Next, create an Analyze format header using
MRIcro's header information
panel. The bare minimum is to specify the
number of voxels in each dimension (the X, Y, and
Z dimensions), the number of bytes per voxel (the
type drop down menu, e.g. 32-bit real type
specifies 4 bytes [a 'byte' is 8 bits] per
voxel), the image offset in bytes (usually zero).
If you are unsure of your image dimensions, read Dave
Clunie's Quick and Dirty Tricks for viewing
medical images.
- If your raw data file contains multiple MRI scans
in the same file, you need to specifiy the number
of 'Volumes'. The size of the raw data in bytes
should be at least X*Y*Z*N*V+O. Where X,Y and Z
are the image dimensions, N is the number of
bytes per voxel, V is the number of volumes and O
is the offset (if the offset is set to a negative
value, each indvidual volume will have its own
offset, so the image size will be
X*Y*Z*N*V*-1*O).
- If you are dealing with raw data, set the
'offset' value in the header information panel to
zero. If your file is in an unknown filetype
instead of raw data, you can usually assume that
the file contains a header at the start of the
image. For example, if your image is a 256x256x1
voxel scan, with 2 bytes per voxel, the raw image
data should be 131072 bytes long - if your file
is 131224 bytes long, set the header offset to
152 (131224 - 131072).
- Save this header by pressing the floppy disk icon
in the header information panel. Give the header
the same name as your image, but using the
extension ".hdr" for your header.
- Now open your image by pressing on the "open
image with displayed header" button (it is
in the header information panel, and shows a
folder with the letters "img").
- Note that most image formats store their data
starting from the upper left corner, storing data
in the same way we read English (left to right,
top to bottom). However, the Analyze format
stores the data starting with the bottom right
(going right to left, bottom to top). Therefore,
you may need to use MRIcro's "Save
as..." function to find the correct
orientation.
- History. Version 0.99 was the first release.
Version 1.0 includes projection views. Version 1.01 can
export regions of interest as 8-bit Analyze format
images. Version 1.02 allows you to create regions of
interest on images with up to 1024x1023 voxels in the XxY
dimension. Version 1.1 added the ability to show
proportionally scaled images, reports the mean intensity
of ROIs. Version 1.11 previews image rotations, and
allows you to navigate projection views by clicking on
the desired location. Version 1.12 can display slice
Z-values in multislice views. Version 1.13 includes free
rotation (oblique slices), data type conversion, and the
ability to transfer regions of interest to images with
different coordinates. Version 1.14 can convert Analyze
images into MRIcro's ROI format. Version 1.15 implements
the ability to yoke projection views. Version 1.16
introduces intensity histograms, the 'hot metal' colours,
the image intensity colourbar and image intensity
autobalancing contrast. Release 1.17 can write to
universal naming convention servers, adds the ability to
turn off graphics acceleration, adds additional features
to the multislice options, and uses less RAM for 16-bit
images. Version 1.18 added XBars and watermarks to all
views, allows adjustment of fonts, allows customized ROI
colours, optionally creates translucent ROIs on
multislices. In version 1.19, the multislice view can
show overlapping brain slices to reveal only the left or
right hemisphere, smoothed ROIs can be saved in Analyze
format. Version 1.20 introduces coronal multislices, the
ability to draw ROIs on sagittal and coronal as well as
transverse slices, and the ability to read and extract
multiple volume Analyze format files. Version 1.21 can read and write DICOM format images.
MRIcro 1.22 can read Genesis format files, the
'File/Export ROI as Analyze image' will now save multiple
overlapping ROIs, right-clicking the ROI information
button will give verbose information about ROI
intensities and Talaraich coordinates, a DICOM-style precise contrast control has been
added, floating point Not-A-Number data is preserved, and
a routine to convert a series of 2D
Genesis or DICOM format scans to a single 3D Analyze
format image. Version 1.23 adds support for ECAT and
Interfile formats, all standard views can be yoked, ROIs
can be intensity filtered (e.g. so that only white or
gray matter is included in the ROI). Version 1.24 added
the ability to open Picker CT and Siemens Magnetom Vision
format, the ability to see the same slice from multiple
images (useful for looking at motion and activity in fMRI
scans) and supports Osiris format color schemes. Version
1.25 can read and convert SPMwin vhd headers, supports
raw binary colour schemes (768 byte color tables with 256
bytes sequentially for red, green, and blue), and has
improved support for 32-bit images. Release 1.26 adds a
magnifier tool and 'smooth' image scaling option. ROI
tools have been added (union, intersection, masking and
subtraction) as described in the tutorial. Version 1.27
allows ECAT image data to be scaled using the 'scaling
factor' and 'calibration' values. When drawing ROIs,
there is now an 'undo' function. Individual slices and
regions can have their ROIs segmented based on image
intensity (as described in the tutorial). Release 1.28
can convert JPEG-compressed (lossless, lossful and XA)
DICOM images to Analyze format, can invert colors, and
can save images as JPEG format. Version 1.29
views/converts Elscint, Par/Rec and Somatom images.
Version 1.30: New installer/uninstaller, Linux
version, SSE support. Release 1.31 adds PNG support and
can save a 2D bitmap graphic for each slice of a 3D
image, hotspots
can be added to coronal or axial slices. Version 1.32 can
display volume
renderings, "Select
MNI/Talairach Coordinates" command in
"View" menu and supports image
overlays (e.g. statistical maps of functional data).
Volume 1.33 adds improved quality surface
rendering, improved statistical overlays
and includes support for VoxBo CUB1 format images.
Version 1.34 includes a better sample MRI scan, improved
BET, and tweaks to the rendering and foreign image
conversion. Version 1.35: includes anatomical
templates , improved importing of
unsupported images , can convert anisotropic
images to be isotropic, opens gzipped Analyze .img
files. Version 1.36 includes image despeckling and
an arcball tool for setting the
viewpoint of rendered images; improves Siemens image
conversion [thanks to Siemens] and speeds up rendering
[thanks to Tom Womack]; improves the quality of surface
renderings; opens confocal images (BioRad and Zeiss
TIFF), cluster size can be set for overlays, basic 3D ROI
drawing, holding the CTRL key down while drawing ROIs
implements a thicker brush, in projection views,
SHIFT+F1/SHIFT+F2 moves up/down coronal slices,
CTL+F1/CTL+F2 moves up/down sagittal slices. Version 1.37
includes a 'Conversion Wizard' ['Import' menu] that can
convert a series of DICOM images to Analyze format in a
single, automated step. Version 1.38 improves contrast
and brigthness settings, using the medical imaging
standard 'window centre' and 'window width' values. Version 1.39 introduces support for NIfTI (.nii, .nii.gz and .hdr/.img files) and support for FSL 3.2. Version 1.40 improves image conversion for newer scanners.
- Additional color schemes: MRIcro can display
images in the "black and white" as well as
"hot metal" color schemes. Additional
Osiris-format color lookup tables (LUTs) can also be
displayed. The Osiris format is a text format, so you can
create
your own custom LUTs. In addition, MRIcro can support
768-byte binary data LUTs (NIH Image, XMedcon format).
MRIcro expects the *.LUT look-up tables to be placed in
the same directory as the MRIcro.exe program. Additional
color schemes are available by downloading the free XMedcon 'extra
stuff' software.
- Platforms, limitations, alternatives: MRIcro
supports the Windows and Linux platforms. Here is a short
list of some freeware Analyze format image
viewers that are currently available for the Windows
PC (for a list of Linux software, see the MRIcro for
Linux web page. A more extensive list of DICOM
viewers for Unix, Macintosh and PCs is available at my DICOM page):
| Viewer |
Platforms |
Formats [Notes] |
| SPM |
Unix/Windows NT |
Analyze [requires Matlab] |
| Slice
Overlay |
Unix/Windows NT |
Analyze [requires Matlab] |
| SPMwin |
Windows |
Analyze |
| ACTIV
2000 |
Windows |
Analyze/DICOM/GE/GIS/PAR/Siemens |
| Medal |
Windows |
Analyze/DICOM |
| etdips |
Windows |
Analyze/DICOM/TIFF |
| Spamalize |
Unix/Windows/Macintosh |
Analyze/GE/TIFF [requires IDL] |
| AMIDE |
Unix/Windows |
Analyze/DICOM/ECAT6/Interfile |
| XMedCon |
Unix/Windows |
Analyze/DICOM/ECAT6/Interfile |
| ezDICOM |
Windows |
Analyze/ECAT/Interfile/Siemens/Picker/GE/DICOM/VoxBo
|
| ImageJ w. Analyze
plugin |
Unix/Windows/Macintosh |
Analyze/DICOM |
| Space |
Windows |
Analyze/VOL [beta release] |
- Linux. A native Linux-native
version of MRIcro is available (MRIcro for Windows can also be used in Linux by using WINE). Alternatively, try the new MRIcron for Linux.
- Macintosh. There are a couple techniques for
running the Windows
version of MRIcro on a Macintosh. Alternatively, try the new MRIcron for Macintosh.
- Converting ROIs. MRIcro stores Regions of Interest using a simple compression technique.I have written a web page that describes tools for converting between the ROI and Analyze or NIfTI formats.
- Updates. The latest versions of the MRIcro
software and help manual are available from www.mricro.com .
 |
















