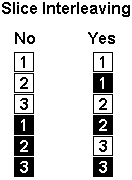 |
MRIcro
tutorial |
This guide gives is a brief description of how you can use
MRIcro and SPM to work with patient scans. For the MRIcro manual
and software, visit the MRIcro home page
(also available at www.icn.ucl.ac.uk/groups/jd/mricro/mricro.html).
I also keep a have a FAQ
that describes some of the more unusual features of MRIcro.
Information about SPM can be found at www.fil.ion.bpmf.ac.uk/spm/,
or at www.mrc-cbu.cam.ac.uk/Imaging/.
This tutorial will describe the following features:
- Download this tutorial
- Viewing images
- Creating regions of
interest (marking and viewing lesions)
- Segmenting a region of
interest
- Linear and nonlinear
normalization functions
- Normalize paitent
scans in stereotactic space (SPM5)
- Normalize paitent
scans in stereotactic space (SPM2)
- Normalizing patient
(or EPI) scans in stereotactic space ( SPM99 )
- Normalizing
patient scans in stereotactic space ( SPMwin )
- Creating multislice
images
- Creating multiple
slices that only show one hemisphere
- Working with multiple
regions of interest
- Yoking images to check
registration
- Displaying a rendered
image of the brain's surface
- Batch processing scanner
images
Downloading
this tutorial
- Windows users can download this tutorial by downloading
the Windows
installer.
- Linux users can download a zip file: shift+click
here. This will copy a compressed version of the
MRIcro web pages ('mritut.zip') to your computer (The
zipped file requires around 450 kb).
- The MRIcro installation include a 1x1x1mm MRI scan. This is
useful for learning how to use MRIcro. The image is named
'ch2bet.img': this is scalp-stripped version of the
average of 27 T1-weighted scans of the same individual as
displayed at www.bic.mni.mcgill.ca/cgi/icbm_view).
A version of this image with the scalp included is also
included with the latest MRIcro releases (this file is
named ch2.img). Note that this MNI image does not
exactly match the Talairach atlas. However, an MNI
atlas is available.
Viewing an image
- Select 'Open Analyze Format hdr+img...'
from the 'File' menu. You will be asked to select the MRI
you wish to view. At the Medical Research Council, the
(normalized) T1 scans have the name 'n'+scan number (e.g.
n12345) while the T2 ('pathological') scans have the name
'p'+ scan number (e.g. p12345).
- The 'slice viewer' panel has three
sliders: the top slider determines the scan slice you
will be shown.
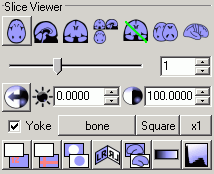 |
Left: The 'Slice
viewer panel'. Controls on this panel allow you
to adjust the appearance of the image you have
opened. |
- You can use the 'black' and 'white'
sliders to adjust the brightness of the image. An
automated brightness can be selected by selecting
'Contrast autobalance' from the 'View' menu. With some
images (those with more than 256 levels of gray),
adjusting the brightness sliders will cause the 'optimise
contrast' button to appear (in the figure above, it is
the black and white circle). Selecting 'Contrast
autobalance' from the 'View' menu usually improves the
image's appearance.
- The bottom of the slice viewer panel
allows you to select the cut you wish to view
(transverse, sagittal, coronal, projection, free rotate
or multiple slice views are available).
- An easy way to view an image is to select
the projection view, and then click on the brain image to
select the coordinates you wish to view.
Creating a region of interest
- Run MRIcro and select 'Open Analyze Format
hdr+img...' from the 'File' menu. Enter the name and path
of the file you wish to view.
- You will probably only want to create a
region of interest on the T1 weighted scan, but you may
want to open up a T2/proton density/T2* image to check
the lesion location. For more information on different
types of MRI scans, visit Joseph P. Hornak's The
Basics of MRI web page. The
Whole Brain Atlas also
has examples of MRI/CT data.
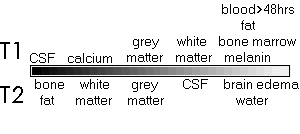
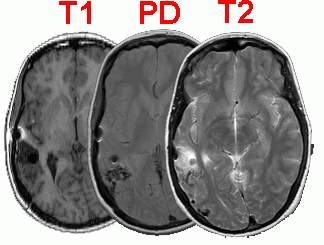 |
Left: T1, proton density and
T2 weighted slices from the same patient. The
'pathological' T2 scan is useful for locating the
lesioned region in the brain. The 'anatomical' T1
scans usually have the best scan resolution, and
are useful for localizing anatomical structures.
The PD scan shows overall hydrogen density per
cubic mm. Note artifacts caused by an aneurysm
clip located directly anterior to the lesion
(black hole, with a white halo in these images).
|
| Right: Relative brightness
levels for different material in T1 and T2 scans.
Notice that lesions are generally bright in T2
scans. |
 |
- Regions of interest can only be drawn on
the transverse, coronal and sagittal slices of the images
(MRIcro versions prior to 1.20 only allow ROIs to be
defined on the transverse slices). In the slice viewer
panel, click on the button that shows a minature axial
view of the brain. If the image does not appear to be
sliced axially when the transverse button is selected
(e.g. the scan looks coronal), then you may want to
rotate the image, as described in the next section.
- Press F3 to select the closed pen (or
click on this tool in the region of interest panel).
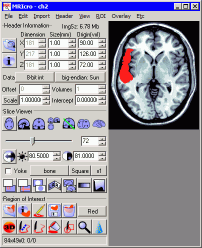 |
Left: Drawing a
region of interest on a scan. |
- Go to the slice you wish to edit (you can
use the F1 and F2 keys to go to the next lower or higher
slice, respectively)
- Draw the region of interest by dragging
the mouse cursor around the border while depressing the
left mouse button.
- Go to the center of the region, an click
the right mouse button to fill the region.
- If you make a mistake, press the icon of
the hand holding an upside down pencil (it is in the
region of interest panel). This will erase your region of
interest from this slice only. You can also shift-click
to delete a region of a ROI. For example,
shift-left-clicking deletes a region of interest beneath
the mouse's movements, while shift-right clicking deletes
a filled-region underneath the mouse's position.
- Repeat steps 5-8 until you have drawn the
lesion on all of the slices.
- Choose 'Save ROI...' from the 'ROI' menu.
It is often useful to give the ROI the same name as the
image file (e.g. if the image file is n12345.img then the
roi should be as n12345.roi).
- Open the next image you wish to view.
Repeat until you have created region of interest
information for all of the images you want to process.
Note: MRIcro 1.36 and later includes
tools for creating intensity-defined 3D regions
of interest.
Segmenting
a region of interest
MRIcro can also be used to filter a region of
interest based on the image intensity of the scan. For example,
consider an fMRI study of auditory cortex. The experimenter has
drawn a region of interest to delineate this region of the
cortex, but is specifically interested if the activity of the
gray matter is modulated with the task. To solve this problem,
the experimenter could filter the ROI of auditory cortex in order
to remove white matter from the analysis. Note that SPM99 has a
series of sophisticated segmenting algorithms that may be more
accurate than MRIcro in many instances. MRIcro can apply an
intensity filter to the entire volume (so the same contrast
levels are applied to all of the slices in the entire image), to
a single slice or to a region within a single slice.
Here is a step-by-step guide for segmenting an
ROI using MRIcro:
- Draw your ROI as described in the previous section.
- Make sure to adjust the contrast of your
image so that the different tissues appear visually
distinctive. For images with more than 8 bits per voxel
you may want to use the precision contrast tool (select
'Precise contrast' from the 'Viewer' menu).
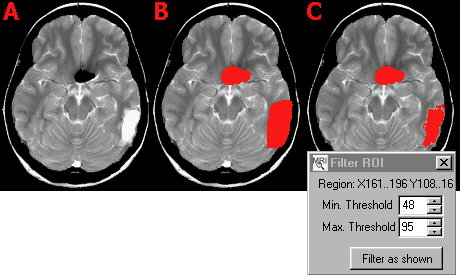
- Select 'Apply intensity filter to
slice/region...' from the 'ROI' menu. You will then be
asked if you want to apply the contrast filter to the
entire slice or a region of the slice. If you want to
apply the filter to only a region, you will be asked to
click on the top-left and bottom-right corners of the
region you wish to filter. In the example below, we want
to apply a different threshold to the dark clip artifact
than the bright lesion. Therefore, we need to select the
appropriate region.
- Adjust the intensity thresholds so that
the tissue you want to preserve is highlighted in green.
For example, when selecting the gray matter, the image
might look like the panel C in the figure below.
- Press the 'Filter as shown' button.
- If you plan to use the ROI with SPM, you
will want to convert the ROI to an Analyze format image.
To do this, choose 'Export ROI as Analyze image...' from
the 'ROI' menu.
Linear
and nonlinear normalization functions
SPM can be used to 'normalize' MRI scans. These
functions reorient the angle, position, size, etc of the scan to
standard stereotaxic space. Normalized brains are much easier to
read, because the slices match those available in published
atlases. In addition, the majority of modern functional imaging
studies normalize their scans to standardized space. Therefore,
if you normalize your patient scans, the resulting scans will be
easy to interpret relative to each other, to atlases and to
functional imaging studies. However, SPM's normalization routines
can be disrupted by a patient's lesions. This is because the
lesioned region is very different from the corresponding region
in the template (that comes from a healthy individual). In
particular, SPM's non-linear
routines are sensitive to lesions, and these
routines will try to eliminate the lesion by compressing that
region (distorting the shape of the brain in the process, see the
figure below). There are two solutions. First, you can normalize
the brain using only the linear
components of the coregistration (e.g. set the
parameter estimation defaults so that there are no nonlinear
iterations and no basis functions). This method tends to do a
fairly decent job of coregistering the brain. If you are using
SPM99, you can take advantage of its 'object masking' feature to
coregister a patient's scans. With this method, the masked region
(e.g. area of a lesion) does not influence the parameter
estimation. The advantage of this method is that SPM can apply
nonlinear basis functions to the image, allowing a better
coregistration. In addition, the lesion does not influence the
linear components of the normalization, allowing a better linear
fit as well. This section describes how to implement this
technique.
 |
Left: Linear, linear+nonlinear, and
masked linear+nonlinear (respectively) normalizations of
the same image to the MNI representative brain (shown on
the far right). Note that without a mask (2nd from left),
the nonlinear components compress the lesion, hiding the
true position and size of the lesion. This example is
from Matthew Brett. |
The table below shows each of the linear
functions (translation, rotation, zoom and shear) applied to
representative input images. Note that all of these functions can
be applied in each of the three dimensions (e.g. rotation can be
applied in the x, y and z coordinates, correcting yaw, pitch and
roll of the input image). With the linear functions, note that
any three points that are colinear in the input image will also
be colinear in the output (though two parallel lines in an input
image may not be parallel after an affine transformation). Also,
all of the 12 linear parameters (translation, zoom, rotation and
affine functions each in the x,y and z dimensions) are computed
based on information from the entire image -therefore these
functions are rarely disrupted by unusual local features found in
MRI scans (e.g. aneurysm clip artifacts or lesions). The final
row in this table shows how a nonlinear basis function can be
used to coregister a brain image. In the representative example,
the input image has a fisheye effect, which can be compensated
for by a simple basis function. Unlike the linear functions,
points that are colinear in the input image may not necessarily
be colinear after nonlinear normalization. Furthermore, nonlinear
functions are more heavily influenced by local image information,
and therefore nonlinear normalization of patient scans may lead
to distortion due to the unusual appearance of the lesion or
aneurysm clips. For this reason, it is important to mask lesions
and clip artifacts when estimating a nonlinear normalization (see
the next section, which describes lesion masking in SPM99).
Normalize patient MRI scans with
MRIcro and SPM5
The best way to normalize images with SPM5 is to use the unified normalization routine (simply press the 'Segment' button in SPM5). This option appears to give better solutions than previous versions of SPM or the standard normalization (which you get if you press SPM5's 'Normalize' button). Further, it does not appear that lesion masking is necessary if you use the unified normaliazation with brain lesions (see Crinion et al. (2007).
Normalize patient MRI scans with
MRIcro and SPM2
This section presents a step-by-step guide for
implementing the technique described by Brett et al. For
a reprint of this article, please contact Chris Rorden.
For references, please cite:
- Brett, M., Leff, A.P., Rorden, C., Ashburner, J.
(2001). Spatial normalization of brain images
with focal lesions using cost function masking.
NeuroImage, 14, 486-500.
|
This section describes how to mask lesions so they do not
disrupt the automated normalization functions of SPM2. Using this
technique, SPM2 can accurately align a patient's scan to
stereotaxic space without causing undue distortion. To complete
this you will need SPM2 beta (or later) and MRIcro version 1.19
or later.
Stage 1: using MRIcro on a Windows PC:
- Open the patients MRI scan and ensure that
the scan is in the correct orientation and you have
marked the anterior commissue (see Stage 1, steps 1-2 of
the section on
normalizing patient scans with SPM99).
- Use MRIcro's Region of Interest tools to
create a ROI that accurately maps the location and extent
of the lesion (see the earlier section on creating ROIs).
- Make a lesion image. With your image and
ROI open, choose 'Export ROI as Analyze image...' from
the 'ROI' menu.Choose 'ROI is 1, background is 0' and
save the image with the prefix 'l' (for 'Lesion', so if
your file is named 'filename.img', you will have a new
image named 'lfilename.img'.
- Make a masking image. With your image and
ROI open, choose 'Export ROI as smoothed Analyze
image...'. Choose a FWHM smooth of 8mm, a 0.001%
threshold and make sure you select 'ROI is 0 [SPM object
mask]'. Save your image with the prefix 'm' for mask. So
if your original filename was 'filename.img', the mask
image should be 'mfilename.img'.
Stage 2: using SPM2 to normalize your image:
Note: you will want to have the
file 'lesionmask.m' in the same folder as SPM2. This simple
script will allow you to quickly normalize images with SPM2. You
may want to read through this script to ensure that it is
accurate for your images: it assumes you are converting
T1-weighted images. In addition, it is assumed that you have 3
Analyze files for each individual, all in the same folder:
'filename.img', 'lfilename.img', 'mfilename.img'. These files
represent the MRI scan, the lesion definition and a smoothed
version of the image that is used as a mask.
contents for
lesionmask.m (click
here
to download; place in spm folder): %red text are comments,
not code
spm_defaults %initialize spm variables
global defaults %initialize
varialbes
dnrm = defaults.normalise; %normalization is based on SPM defaults
dnrm.write.vox = [1 1 1]; %set resilcing of high resolution images for 1mm
isotropic
dnrm.estimate.wtsrc = 1; %use mask image for normalization
dnrm.estimate.weight =
fullfile(spm('Dir'),'apriori','brainmask.mnc'); %use a mask for the template
image: we want to align shape of brain, not the scalp
dnrm.write.bb = [[-91 -125 -71];[89 91 109]]; %set bounding box to match
brodmann and AAL maps that come with MRIcro
VG0 = spm_vol(fullfile(spm('Dir'),'templates','T1.mnc'));%match scans to the T1-weighted
template
V = spm_vol(spm_get(Inf,'*.IMAGE','Select images')); %have the user select image
for i=1:length(V), %repeat
for each image the user selected
[pth,nam,ext] = fileparts(V(i).fname);%segment file path/file name and
extension
wtsrcName = fullfile(pth,['m' nam ext]); %the mask image has the prefix
'm'
VF = V(i); %select
the MRI scan
matname = [spm_str_manip(V(i).fname,'sd')
'_sn.mat']; %name
for file that will store transformation matrix
prm =
spm_normalise(VG0,VF,matname,dnrm.estimate.weight,wtsrcName
,dnrm.estimate); %normalize
an image based on our preferences
spm_write_sn(V(i),prm,dnrm.write); %reslice the image to new
coordinates and save to disk
LesionName = fullfile(pth,['l' nam ext]); %the lesion image has the prefix
'l'
spm_write_sn(LesionName,prm,dnrm.write); %reslice the lesion to new
coordinates and save to disk
end; %repeat for
each image |
- Open Matlab and type 'lesionmask' into the command
window.
- You will be asked to select your images. Only select the
MRI scans (e.g. filename.img), you should not
select the mask (mfilename.img) and lesion
(lfilename.img) images. Note you can select multiple
files to batch convert images.
- SPM2 will then normalize images, the output will be a
normalized version of the MRI scan ('wfilename.img') and
a normalized copy of the lesion ('wlfilename.img').
Stage 3: using MRIcro on a Windows PC:
- Save the lesion in MRIcro's compressed ROI
format. Open the lesion defintion scan (e.g.
'wlfilename.img') select 'Export Analyze image as ROI...'
('ROI' menu). You will be asked to set a minimum
threshold for accepting a region as a lesion: you should
set this value to 50 (50% of the maximum image
intensity). Give this the same name as the patients
normalized image (the ROI for image 'wfilename.img'
should be 'wfilename.roi'.
- Open the normalized patient image with the
'Open image...' command ('File' menu). This should load
both the normalized image and (if you have successfully
completed the previous step) the normalized ROI. Notice
that the volume of the lesion is displayed in the region
of interest panel. Make sure to save your ROI. You can
now view the lesion from any view and get a good idea of
the size/position of the lesion (all in standardized
stereotactic space).
Normalize
patient MRI scans with MRIcro and SPM99
This section presents a step-by-step guide for
implementing the technique described by Brett et al. For
a reprint of this article, please contact Chris Rorden.
For references, please cite:
- Brett, M., Leff, A.P., Rorden, C., Ashburner, J.
(2001). Spatial normalization of brain images
with focal lesions using cost function masking.
NeuroImage, 14, 486-500.
|
This section describes how to mask lesions so they do not
disrupt the automated normalization functions of SPM. This
technique is also useful for EPI fMRI images from healthy
individuals (where EPI artifacts can also disrupt normalization).
The MRC-CBU has a page
devoted to EPI masking. Souheil
J. Inati has also posted a useful technique for normalizing
EPI scans with SPM.
SPM99 can compute normalize patient scans to a
stereotaxi standard template using both linear and nonlinear
functions. Allowing SPM to apply nonlinear transformations will
generally result in a more accurate normalization. However, with
patient scans, nonlinear functions can effectively squash the
region of a lesion (as it has no equivalent in the healthy
template brain), causing considerable distortion of the brain.
This section describes how to create and apply a lesion mask with
SPM99, which allows accurate linear and nonlinear normalization
of scans that have large lesions.
You will require SPM 99 beta (or later) and
MRIcro version 1.19 or later (to get the latest version of
MRIcro, go to the MRIcro
home page).
Stage 1: using MRIcro on a Windows PC:
- Open your scan and check the image
orientation and the amount of neck presented in the
image: when the slice viewer panel is set to display
transverse slices, you should see transverse slices with
the anterior portion of the brain (and the nose) toward
the top of the screen and the posterior regions of the
brain toward the bottom of your computer screen.
Furthermore, the amount of neck displayed in your image
should roughly match the amount of spinal cord shown in
the template image. If your image is not in the correct
format, choose the 'Save
as...[rotate/format/clip/4D->3D]' from the 'File'
menu. A window will appear which allows you to describe
the current orientation of the brain and the amount of
the spinal cord you wish to clip from the image. The
preview button allows you to ensure that you have
selected the correct orientation. Once you have correctly
described the image orientation, press the 'save Intel'
(or 'Save Sun', both SPMwin and MRIcro can read either
format). Advanced
note: if all of your scans are in the same non-transverse
format (e.g. coronal), you can set SPM96/SPM99 [but not
SPMwin] to automatically rotate the scans for you by
setting the 'sptl_Ornt' parameter of your spm_defaults.m
file. For more details, search the SPM archives for 'sptl_Ornt'. Another option
is to reorient the images using SPM99's 'display'
feature.
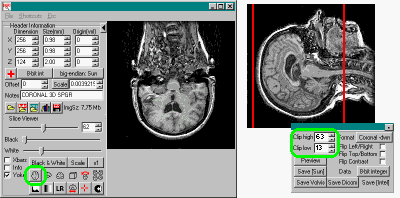 |
Left: The left panel shows a
coronal format scan. Note that when the
transverse view button (highlighted in green)
is selected, the scan appears to
be coronal. The right panel shows MRIcro's scan
rotation panel, with the format for the scan set
to coronal. Note that the rotation panel has
'clip low' and 'clip high' settings, which allow
you to interactively shave slices off of the
image. In this example, I want to remove some of
the excess spinal cord, so that this region will
not influcence SPM. Note that the sagittal view
on the right shows much more spinal cord than the
template brain (shown in the projection view,
below). |
- Make sure that the 'origin' fields
correctly designates the location of the Anterior
Commissure (see the diagrams below to get an idea of
where the AC is located, or read Matthew Brett's AC-PC web page). To find the AC, click on the projection views
button (it looks like a cube) and make sure that the
'XBars' command is selected. Then, use the mouse to click
on the location of the AC (in the example below, the AC
is located near 46 64 37). Once the image cross-bars are
positioned at the AC, input the X Y and Z coordinates
listed in the 'slice viewer' panel into the origin fields
of the 'header information' panel. You should then save
the header (press the floppy disk icon located in the
header information panel).
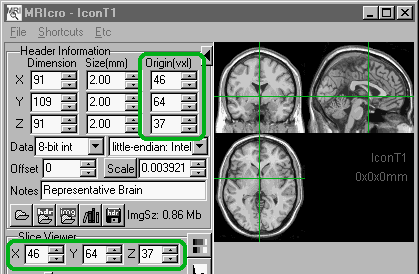 |
Left: The location
of the anterior commissure (AC) on the Montreal
Neurological Institute's representative brain.
The location of the crossbars (which are now at
the AC) is given at the top of the slice viewer
panel (lower
left) . These values should
be placed in the 'Origin' fields (upper left) of the header information panel in order
to tell SPM the location of the AC. |
- Making the lesion definition image. Open
the source image with MRIcro and create a region of
interest (see the earlier section on creating ROIs) that includes the region of the lesion only.
Using MRIcro's 'Save ROI as Analyze image...' command
(from the 'ROI' menu) to convert the lesion into a format
that SPM can read. As a convention, I usually add
the 'L' prefix to these lesion definition files, so the
lesion for image '123.img' should be 'L123.img'. Advanced note: If you draw the
lesion in one plane only (e.g. drawing on transverse
slices only) your ROI will usually have discontinuous
stepping artifacts in the the dimension orthogonal to the
plane you are drawing in (e.g. when drawing the ROI on
transverse scans, you are marking images in the X and Y
dimensions, and the Z dimension may having stepping
artifacts). These irregularities will look unusual when
SPM normalizes the lesion into stereotaxic space. One
solution is to use MRIcro to save smooth the lesion
before normalization. To do this, choose the 'Export ROI
as smoothed Analyze image' function [instead of the
unsmoothed function] and select a narrow smoothing band
[4mm] will a 50% cutoff [Threshold=0.5], and make sure
that the dropdown box selects 'ROI is 1 [reslice ROI].
Press save, and give the file a suitable filename for a
lesion definition (e.g. my convention is to add the 'L'
prefix to the file name). Saving the lesion definition
with these smoothing values will create an image file
that is no larger than the original lesion but does not
have any specific stepping artifacts.
- Make the mask image -if the MRI scan
includes any regions of unusual appearance besides the
lesion (e.g. any clip artifacts), add these to your
MRIcro ROI. Essentially, you want to mask out any unusual
regions of the brain that do not have an equivalent on
the template image and therefore can throw off SPM's
normalization routines. Then save this as an image mask
by choosing the 'Export ROI as smoothed Analyze
image' function. This will save the region of interest as
an Analyze format image. You will use this as the mask in
SPM. As a convention, I usually add the 'm' prefix to
these mask files, so the mask for image '123.img' should
be 'm123.img'. Advanced note: This
window has a number of options for controlling the amount
of smoothing, the full width half maximum (FWHM)
describes the width of the smoothing function, which
should be the same as you are using in SPM (often 8mm).
The threshold sets the mask's sensitivity to the presence
of an ROI: 0.001 is standard. In addition, you can set
how the filter deals with the edge of an image -you will
probably want to keep the setting at 'Adjust sides in
Z-plane only [SPM]', which precisely mimics SPM's
behavior (and will show the same edge boundary artifacts
as SPM). Finally, make sure that the final drop down box
is set for 'ROI is 0 [SPM object mask].
Stage 2: Using SPM99 on a UNIX machine or
with Windows NT:
- Launch SPM99. This will vary depending on your system,
usually you start Matlab and then type 'SPM' in the
Matlab command window.
- Press the 'PET/SPECT' button (or the 'fMRI' button, it
does not matter for image normalization).
- Check the settings by pressing the 'Defaults' button and
choosing "defaults/spatial normalization/parameter
estimation"to make sure that the "masked object
brain"setting is set to "true".
- Use the 'Display' button to check that the image
horizontal plane is roughly that of the AC-PC line (see
the figure above, you may need to adjust the 'pitch'
setting). If you change the image orientation, make sure
to save the settings by pressing 'reorient images'.
- You are now ready to normalize the images.Press the
'Normalize' button and choose the 'Determine parameters
and write normalized"function.
- You will be asked to locate four sets of images:
- The image you wish to get the parameters from
(the patient's scan, e.g. 123.hdr)
- The mask file (e.g. the mask you made from the
patients ROI file, e.g. m123.hdr).
- The images you wish to write these parameters to
-the patient's scan (e.g. 123.hdr)
-the lesion definition file (e.g. L123.hdr)
- The template image (e.g. look in SPM's template
folder and choose T1.img)
- SPM will now normalize your brain. The output files will
be named with the prefix 'n' (e.g. if your patient's
brain is 123.hdr', the output will be 'n123.hdr', and if
the lesion definition is 'L123.hdr', the output will be
'nL123.hdr').
Stage 3: using MRIcro on a Windows PC:
- Save the lesion in MRIcro's compressed ROI
format. Open the lesion defintion scan (e.g. 'nL123.hdr')
select 'Export Analyze image as ROI...' ('ROI' menu). You
will be asked to set a threshold for accepting a region
as a lesion: you should set this value to 50 (50% of the
maximum image intensity). Give this the same name as the
patients normalized image (the ROI for image 'n123.hdr'
should be 'n123.roi'.
- Open the normalized patient image with
'Open Analyze format .hdr + .img' ('File' menu). This
should load both the normalized image and (if you have
successfully completed the previous step) the normalized
ROI. Notice that the volume of the lesion is displayed in
the region of interest panel. Make sure to save your ROI.
You can now view the lesion from any view and get a good
idea of the size/position of the lesion (all in
standardized stereotactic space).
Normalize patient MRI scans with
MRIcro and SPMwin
SPM for windows (SPMwin) is a freely
available standalone implementation of
SPM96. Unlike other versions of SPM, SPMwin does not require
Matlab. Therefore the software is completely free and many users
find the user interface friendlier than Matlab versions of SPM.
However, the current release of SPMwin does not implement SPM99's
ability to create object masks for normalization of patient scans
(see the previous section for details). However, for the majority
of patient scans, SPMwin can do a reasonable job of coregistration,
with the caveat that the normalization should only include
linear transformations. The nonlinear transformations will often distort
patient scans in an attempt to eliminate the region of the
lesion. See the section
on linear and nonlinear transformationsfor
more details on the advantages and disadvantages of applying
nonlinear transformations to patient scans.
You will require SPMwin and MRIcro version 1.19
or later (to get the latest version of MRIcro, go to the MRIcro
home page)
Stage 1: using MRIcro on a Windows PC:
- Open your scan and check the image
orientation and the amount of neck presented in the image. The
previous section explains this step in detail .
- Make sure that the 'origin' fields
correctly designate the location of the Anterior
Commissure. The
previous section explains this step in detail .
Stage 2: Using SPMwin on a Windows PC:
- Run SPMwin. From the 'File' menu choose the
'new/normalization' command. Once you have told the
computer how many scans you wish to normalize, you will
see a window named 'spatial normalization' with the icon
of a small book. Double click on the icon, you will see
four small icons labelled 'template','image to estimate
parameters' and 'image to normalize'.
- Double click on the template icon. Make sure that the
listed template is the correct one for your patient's
scan (e.g. if you are normalizing a T1 scan, make sure
that the template is 'T1.hdr'). If it is incorrect, click
on the filename and press delete, then drag and drop the
correct template header to SPMwin's template icon.
- Drag and drop the patient scan's header file to both the
'image to estimate parameters' and 'image to normalize'
icons.
- You may want to check the settings by choosing the
'settings' command from the 'calculate' menu. If so, make
sure that 'sync' interpolation is selected in the
'general' tab. Makesure that the 'Number of nonlinear
iterations' and 'Number of basis functions' are both set
to zero (parameter estimation tab). Also check the
reslicing dimensions in the 'reslicing' tab.
- Choose 'start' from the calculate menu. When the
normalization is complete a new image file will be
created, which will have the prefix 'n' (e.g. if your
patient's scan had the name '12345.img', the normalized
scan will be named 'n12345.img').
Stage 3: using MRIcro on a Windows PC:
- Open the normalized scan (e.g. n12345.vhd
and n12345.img). Press the floppy disk icon in the header
informtion window to create an Analyze format header
(n12345.hdr).
- Create region of interest that marks the
region of the lesion (as described in the 'Create ROI'
section). This region of interest should only include
the regions of the lesion. Notice that the volume of the
lesion is displayed in the region of interest panel. Make
sure to save your ROI. You can now view the lesion from
any view and get a good idea of the size/position of the
lesion (all in standardized stereotactic space).
Creating
multislice views
MRIcro allows you to quickly create 'multislice' images that
show a number of preselected cuts from an scan. This can be
useful for presenting similar slices from a number of patients.
This section describes how to generate a multislice view.
- Open the image you wish to view by choosing "Open
analyze format hdr+img". If you want to superimpose
a region of interest [ROI, e.g. an outline of a lesion]
there are two options. First, if the ROI has the same
name as your image (e.g. the image is named
"mr1.img" and the ROI is "mr1.roi",
the ROI will have been opened automatically when you
loaded the image. If the ROI has a different name, or if
you want to superimpose multiple ROIs, select "Open
ROIs..." from the 'ROI' menu. A dialog box will
appear and you can control+click on each ROI you wish to
open [e.g. for the tutorial, you can open
"a.roi","b.roi" and "c.roi"
simultaneously].
- Open the Etc menu's 'Option' window and select the slices
that you want to view (these values
are shown outlined by yellow in the figure below).
You can display up to twelve slices, but if you wish to
show fewer slices enter a zero into the unused fields.
For this stage you must enter the slice number rather
than the Talairach position (e.g. in the image that comes
with this tutorial, Z of 0mm is on the 37th slice, so to
show Z=0mm enter the number 37). An easy way to find the
corresponding slice number is to show the projection view
of an image. You can then adjust the slice number (using
the boxes in the slice viewer panel) and see the
corresponding Talairach coordinate (if the slice viewer's
'info' button is selected).
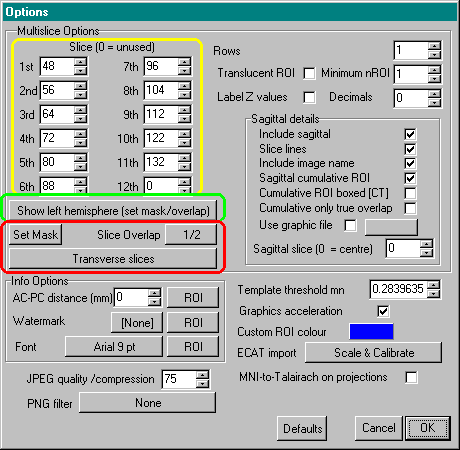
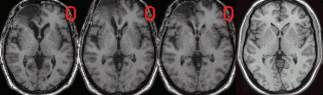
- I typically display axial slices 48, 56, 64, 72, 80, 88,
96, 104, 112, 122, and 132 (for normalized images with
1mm slices, like the image that is downloaded with this
tutorial). These slices correspond to Talairach slices
-24, -16, -8, 0, 8, 16, 24, 32, 40, 50, 60mm, which are
common transverse slices.
- The options window has a number of additional settings
that control the multislice view. Once you have selected
your desired settings, press the 'OK' button. (MRIcro
will remember your preferred settings, so you usually
only have to set these up once). There are a number of
options that you can choose:
- Select whether you want to view Transverse or
Coronal slices. I usually show tranverse views,
but for some images (e.g. showing hippocampal
lesions), the coronal view may be more
appropriate.
- Select whether you want to see a sagittal view.
- Select from the various options for the sagittal
view.
- To generate a multislice view, click the multislice view
in the Slice Viewer panel. You should see a multislice
image similar to the one displayed below.
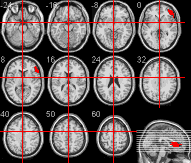
Creating multiple slices that only
show one hemisphere
Recent versions of MRIcro have made this a bit easier and more
powerful. This tutorial describes features in MRIcro 1.35 and later.
- Select 'Open Analyze Format hdr+img...'
from the 'File' menu. You will be asked to select the MRI
you wish to view.
- Open the Etc menu's 'Option' window and make sure that
multislice options are set for 'Show right hemisphere' (this box is outlined in green in the
diagram of the Options window shown in the previous section) , or 'Show left hemisphere' if
you wish to show the left hemisphere. You can also set
the other multislice options by setting the approriate
check boxes and values.
- Set how whether you want to see coronal or transverse
slices, and set how much overlap you want: you can values
between 1/2 (the entire left side will be hidden) to 1/5
(only a small chunk of the left side will be hidden. These controls are outlined in red in the
diagram of the Options window shown in the previous section)
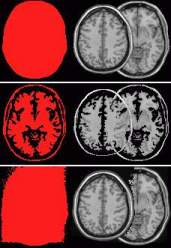
- Press the 'Set Mask' button to interactively adjust the
border threshold (also outlined in
red in the diagram of the Options window shown in the
previous section). A dialog box appears that
allows you to set the boundary threshold for your image.
As you adjust the threshold image, portions of your image
that are above the threshold will appear as the region of
interest colour (red in the left
images on the figure below) . Adjust the threshold
so that the skull defines a boundary for the brain (top panel in the image below) .If
the threshold is set too high (middle
panel below) or too low (bottom
panel below) , the resulting multislice will show
mask artifacts (the right side of
each panel shows the corresponding multislice for each
mask setting).
- When you are happy with your settings, close the
threshold window and close the Options window.
- Press the multislice button in the slice viewer panel (it
has an icon with multiple brain images).
- If you are happy with the result, you can use the 'Save
as picture' command to save the image in a standard
graphic format of your choice.
- WARNING : If you have
set the 'translucent ROIs' checkbox in the Option window,
you should ensure that all regions of interest exist only
in the hemisphere that is to be displayed. Translucent
regions of interest (and for advanced users, functional
imaging hotspots) are added after MRIcro has overlaid the
scans, and ROIs in the hidden hemifield can appear on the
visible hemisphere of the incorrect slice.
Working
with multiple ROIs
It is often useful to display overlapping regions of interest
(ROIs) from different individuals. For example, after you have
normalized the brain scans from several neurological patients, it
can be useful to explore the regions of mutual involvement. There
are a couple of important issues to bear in mind when combining
multiple ROIs. First of all, all of the ROIs need to be from
spatially coregistered images - i.e. the ROIs must have the same
voxel size and image dimensions. Consider if we want to examine
the ROIs from patients A and B. These two patients are shown in
the picture below as 'A' and 'B'. The first thing we might want
to generate is a density plot, that would should us which regions
are mutually involved.This is represented as A+B in the figure
below. To do this, you can conduct the following steps:
- open a template image that has the same spatial
dimensions as the patient's images.
- Open the ROIs using the 'Open ROI[s]...' option from the
ROI menu. Select multiple ROIs by control+clicking on the
different filenames.
- MRIcro now shows the overlayed lesions, as illustrated by
A+B below.
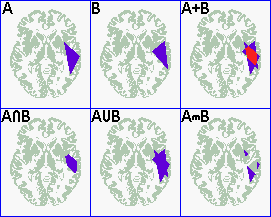
However, it can also be useful to apply boolean (logical)
operators to the ROIs. MRIcro includes three logical ROI
operators - intersection, union and subtraction. Each of these
commands is found in the "ROI comparisons" submenu of
the "ROI" menu. For example, we could find the region
that is involved in two groups of patients. In this case we could
compute the INTERSECTION of the two data sets (a logical
"AND"): Only voxels which are present in ALL the ROIs
will be present in the output ROI. Another reasonable comparison
would be to examine the UNION of two data sets (a logical
"OR"): voxels present in ANY of the ROIs will be
included in the output ROI (AUB in the figure above). A final
useful comparison is MASK, where MRIcro identifies the
region that is unique to A (AmB in the figure above). Note that
in the sample above only two ROIs were compared. In practice, you
can apply the comparisons to many ROIs simultaneously
(control+click on the file names to select multiple ROIs).
The logic and power of subtraction analysis is
described in Karnath et al. For a reprint of this
article, please contact Chris Rorden.
For references, please cite:
- Karnath, H.-O., Himmelbach, M., Rorden, C.
(2002). The subcortical anatomy of human spatial
neglect: putamen, caudate nucleus and pulvinar.
Brain, 125, 350-360.
|
Lesion density plots (A+B in the figure above) have become a
common tool method for attempting to infer the functional role of
an anatomical region. However, you should be careful with the
inferences you draw from density plots. One problem is that the
size and extent of naturally occurring lesions is typically
determined by vasculature (for strokes), and susceptibility to
sheer and impact (for traumatic brain injury) rather than
function. Therefore, simply overlapping lesions from patients who
show a specific deficit will often highlight regions involved
with the function as well as regions that are simply more
susceptible to damage. One potential solution is to directly
compare patients with a specific deficit to control patients with
a SUBTRACTION analysis. Regions that are susceptible to damage
will be occur with similar frequency in both groups, and be
effectively cancelled out. On the other hand, regions that are
required for the target function will tend to be damaged in the
target patients but not in the control patients. In any case,
note that naturally occurring lesions vary greatly in the
location and extent of the damage, adding a huge amount of
variability into an analysis. This large variance substantially
reduces the power of anatomical studies. One solution is to
conduct analysis on a large number of patients. A second solution
is to restrict analysis to patients with focal lesions.
The "ROI comparisons" submenu also allows you to
compare SUBTRACTIONs between two different groups of
patients. This allows you to identify if certain regions are more
common for one pathology than another. To implement this command,
you should first open your template image. Then choose the
"Subtract ROIs" command. You will then be asked to
select the positive ROIs. Next you will be asked to select the
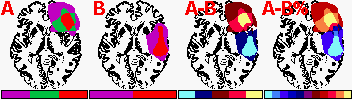
negative ROIs. The figure below illustrates two groups of
patients (A and B). The right panels show the subtraction between
these two groups (A-B, A-B%). You can either subtract the
absolute numbers or relative percentages (the default setting).
Select 'ROI density colorbar' from the ROI menu to see a legend.
The percentage subtraction shows 11 levels of ROI: each bar
represents 20% increments - so the lightest blue represents
81..100% B. The middle purple percentage bar designates regions
where there is an identical percent of A and B (0%). Comparisons
are most meaningful when both groups are the same size.
Yoking
images to check registration
SPM99 includes a feature named 'check reg' that allows you to
view the same Talairach coordinates for two Analyze format
images. This function is useful for testing the normalization of
scans to a template. MRIcro's yoking feature is conceptually
identical, but allows you to check the coregistration of two or
more images simultaneously. Viewing more than two images
allows one to check the normalization of several images to a
template, or to compare to different normalization techniques
with each other.
To view yoked images, launch one copy of MRIcro for each image
you wish to view. Load the images and make sure that the 'Yoke'
checkbox is selected. Once this is done, the images will all show
corresponding slices -irregardless if the images are sliced to
different dimensions (e.g. one image shows 1mm slices, whil the
other shows 2mm slices) or whether the images are cropped
differently (e.g. one image showing more of the skull). To move
to a different position, click on the desired location in any of
the images -the other images will be updated to match your
selection. It is generally useful to shrink the toolpanel and
optimize the formsize (in the 'Viewer' menu) when viewing
multiple images, allowing you to maximize the available screen
space.
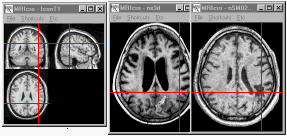
In the example below, three images are yoked simultaneously
-showing (from left to right) a template image and the scans from
two neurological patients. The two left most scans are from the
same patient -notice that the nonlinear functions of the
normalization have compressed the lesion in the rightmost scan
(as described in the normalization
section). Clicking on any position in any of the three scans will
show the corresponding location in all three scans. Note that in
this example the images are cropped differently -with the
normalized images showing less of the skull than the template.
Despite this, the yoking finds the corresponding location in all
three images.
Displaying
a rendered image of the brain's surface.
I now have a web
page devoted to volume rendering with MRIcro.
Batch
processing scanner images.
Note, there are new tools that can convert your images directly to NIfTI format. You can find a listing at my dcm2nii website
Most MRI systems export scans as in either DICOM or a
proprietary format. Unfortunately, SPM and many scientific
packages require Analyze format files. MRIcro can convert many
common medical image formats to the Analyze format file. For a
list of the formats supported by MRIcro and other free converters
see MRIcro's
home page.
This section describes how to stack a series of two
dimensional images to create three dimensional volumes. Most fMRI
studies will generate a vast number of scans, and MRIcro is
designed to ease the conversion of these scans for processing
with SPM or other brain imaging packages.
The batch processing panel is the most complicated feature of
MRIcro for the user. Unfortunately, different scanners use very
different methods for saving images. MRIcro has been designed to
work in many different situations, and this flexibility has meant
that the interface is rather complex. However, images from a
single scanner are virtually always in the same general format,
so once you have tuned MRIcro's conversion the first time,
subsequent conversions should be simple.
As an example, consider a series of DICOM format file's named
a11.dcm...a31.dcm that contain slices from two brain volumes.
Each volume is composed of three slices. Furthermore, only every
fourth is relevant (e.g. the other scans were not T1 images), so
the relevant slices are image a11, a15, a19, a23, a27 and a31. In
addition, the orientation of these scans does not correspond with
the standard SPM axial slices.
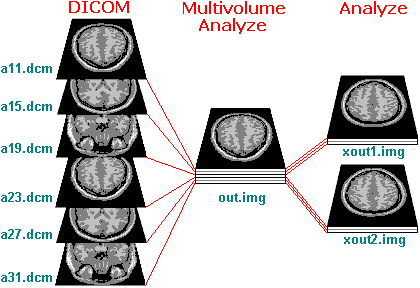
The left column in the figure above shows the six DICOM images
in this example. We will process the images in two steps. First,
we will convert all of the slices to a single multivolume Analyze
format image (that SPM can not use). Next, we will rotate the
images to the correct orientation and convert each volume to a
separate Analyze format file (that SPM can read).
Here is the sequence of steps to batch process these images:
- Launch MRIcro and select 'Convert
DICOM/Genesis/Interfile/Siemens/Picker to Analyze' from
the 'Import' menu.
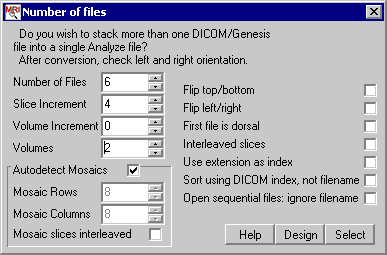
- A new window appears (as shown above). For our example we
will want to adjust the values to match our data set:
- Number of files: Total number of slices to be
converted (in our case, 6 slices).
- Increment: what is the numerical spacing between
successive images (in our case, this should be 4,
as every fourth scan is the T1).
- Volumes: How many brain volumes are we converting
(in our example there are two brain volumes).
- First file is dorsal: Select this to reverse the
slice order (e.g. the slice order is dorsal to
ventral). In our example, slice order is ventral
to dorsal (the first slice [a11] is ventral), so
we will leave this button unchecked.
- Interleaved slices: Check this if the slices from
the different volumes are interleaved, rather
than sequential. In our example, the volumes are
sequentially stored (the first slices [a11, a15,
a19] belong to the first volume followed by the
slices for the next volume [a23, a27, a31]). If
the slices for the first volume were a11, a19,
a27 and slices from the second volume were a15,
a23, a31 we would want to select this checkbox.
- Use extension as index: select this if the image
index number is in the extension, e.g.
a.11,a.15,...a.31. On our files, the extension
(.dcm) is not the index, so we leave this option
unchecked.
- Sort Using DICOM index, not filename: Some
scanners name slices based somewhat randomly
(e.g. saving the files as file1.dcm,
file6.dcm,file2.dcm,file5.dcm,file3.dcm,file4.dcm).
These scanners often save the true sequence in
the DICOM header (group/element 0020h/0013h).
This option will reorder the slices based on
information in the DICOM header. Our files are
indexed correctly by the filename, so we can
leave this unchecked.
- Open sequential files: ignore filename:
Unfortunately, some scanners save images with a
very complicated file naming system that MRIcro
will not be aware of (e.g.
fileA-1,fileB-2,fileC-3,etc). In this case, I
suggest that you write a simple program to rename
the files to a numerical sequence that MRIcro can
interpret (e.g. file1,file2,file3). However, as a
last resort you can have MRIcro ignore the
filename altogether and open sequential files in
a folder. Important: Before using this
option, place the image files you want to convert
into a folder that has no other files of any type
in it. This option may or may not work- your
images may end up in a scrambled order. It works
best best when used in conjunction with the
"Sort Using DICOM Index" option. In our
example, we will [thankfully] leave this option
unchecked.
- Press the 'Design' button to check that you have
correctly described the slice order. A window appears
that shows the slice order, indexed from one. In our
example, the design window will list:
Volume 1: 1, 5, 9
Volume 2: 13, 17, 21
This accurately describes our data with the first volume
storing the 1st, 5th and 9th slices (a11, a15, a19) and
the second volume storing the 13th, 17th and 21st slices
(a23, a27, a31).
- Once you have described the slices, choose 'Select' and
find the first DICOM (or
NEMA, Genesis) file in the series (i.e. the slice with
the lowest numerical value, in our example this is
a11.dcm). The file names must end with the numberical
index (a11.dcm is valid, 11a.dcm is not). Optionally, the
file index may be padded by zeros (e.g. file008.dcm,
file009.dcm, file010.dcm is valid, as is file8.dcm,
file9.dcm, file10.dcm).
- Give a name for the output Analyze format image (e.g.
lets call it 'out.img').
- MRIcro should now process all of the slices and create a
multivolume Analyze format image. If successful, you can
close the DICOM conversion window.
- You will now be able to open the image by selecting 'Open
analyze img+hdr...' from the 'File' menu. You can only
view volumes one at a time, so MRIcro will ask you to
name the volume you want to view.
- You should now check the header information panel and
make sure all of the information is correct.
- You may want to adjust the image intensity
value shown in the header information panel.
SPM will use this scale for determining the
signal level. This is especially crucial for
SPM99b (the final release of SPM99 should do
this automatically). See the SPM99 normalization section
of this tutorial for details on how to
automatically adjust the image intensity.
- Check to make sure that the slice thickness
(Z size [mm]) is correct.
- To extract the individual brain volumes, select 'Save
as...' from the 'File' menu. A new window appears. You
may want to adjust the orientation by selecting the
'format' and checking the results with the preview. In my
example, the images need to be rotated 180 degrees in the
Z plane, so the format needs to be set to 'axial down'.
Click the 'Save Intel' button to extract and rotate all
of the volumes. You will be asked to give the output a
name, for example if you name the output 'xout.img', the
files will be called 'xout1.img', 'xout2.img'...etc.
- You can now delete the multivolume image if you wish (in
our example, out.img).
In our example, each DICOM file contained one image.
Furthermore, the two volumes were stored sequentially rather than
being stored with 'interleaved slices' (that is, all the slices
for the first volume had lower numbers than the scans for the
second volume). Note that the design window allows you to adjust
these features.
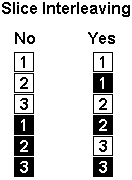 |
Interleaving: In the figure on the left,
slice interleaving is shown on the right panel. In our
example, the volumes were saved sequentially (e.g. all of
the first volume's slices had lower indexes [a11,
a15,a19] than the second volume [a23, a27, a31]).
However, some scanners save volumes in an 'interleaved'
format. Selecting the 'slice interleaving' option will
tell MRIcro that the slices are interleaved (e.g. the
first volume is a11, a19, a27, and the second volume is
a15, a23, a31).
|
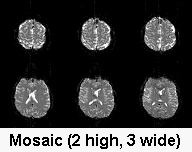
| Mosaics: The NEMA format only specifies
image size in two dimensions, so 3D volumes are stored as
'mosaics', which show various cuts on the same plane. The
figure aboveshows a 2x3 NEMA mosaic. One way to test
whether your images are a mosaic is to select 'Open
foreign' from the 'Import' menu. If you see multiple
slices simultaneously, you are dealing with a mosaic.
MRIcro should automatically detect and convert Siemens
images if you click the 'Autodetect Mosaics' checkbox
(though see Interleave note below). Otherwise, you will
have to manually set the mosaic height and width when
converting the images to Analyze format. Note that the
more recent DICOM format does describe images in three
dimensions, so quick opening these files will show only
one slice per plane (and for conversion purposes, you
should set the mosaic height/width both to 1).
|
 |
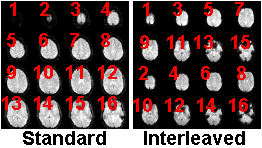 |
Interleaved Mosaics: Most mosaic files store
slices in the same order we read English: starting from
the top left and reading from left to right. However,
some Siemens scanners create mosaics where the slice
order is interleaved within the mosaic. If you select
'mosaic slice interleave', MRIcro will try to correctly
read the order of Siemens mosaics. Note that the
'Autodetect Mosaics' checkbox will NOT detect slice
interleaving for Siemens Vision mosaics, as some fMRI
labs have created and distributed acquisition protocols
that incorrectly set up the image header. Therefore,
Vision users must always manually determine if the mosaic
images are interleaved.
|